Scalable method to image protein levels and changes developed
Posted: 18 November 2020 | Victoria Rees (Drug Target Review) | No comments yet
Using a new CRISPR-Cas9 tagging strategy, researchers have developed a method that enables the imaging of hundreds of proteins in parallel.


Researchers have developed a highly scalable method that allows for the imaging of hundreds of proteins in parallel to monitor the changes of their levels and localisation in the cell. The technique was created at CeMM, the Research Center for Molecular Medicine of the Austrian Academy of Sciences.
According to the team, until now, scientists have typically studied the changes of proteins and their roles in the cell by using a fluorescent tag to label and follow one protein at a time. For this method, a green fluorescent protein (GFP) is fused to the end of a certain protein to be studied. This fusion is then expressed in the cell and through fluorescence microscopy researchers can observe the cells with the labelled protein. This permits the study of many changes, like different drug doses in a time resolved manner for a single protein. However, this approach limits the number of proteins that can be studied and precludes unbiased discovery approaches.
Andreas Reicher and Anna Koren from CeMM Principal Investigator Stefan Kubicek’s group developed a novel strategy which allows scientists to observe and characterise those changes in a very high number of proteins in parallel. This method can be used to not only describe and better understand the effects of certain known drugs in the cells, but also to discover new drug treatments that work by affecting and modulating the protein levels.
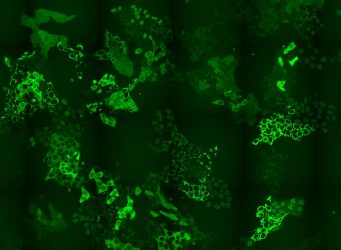

Cell pool expressing hundreds of different GFP-fusion proteins [credit: Andreas Reicher/CeMM].
The researchers have designed a way to overcome a bottle-neck in CRISPR-Cas9-based intron tagging – the need to develop methods that can be used on the whole proteome and not just one protein at a time. The team generated cell pools containing hundreds of tagged proteins and in each cell a different protein was labelled with GFP. These cell pools were exposed to a PROTAC chemical degrader of BRD4, a transcriptional regulator that plays a key role during embryogenesis and cancer development. The researchers then used time-lapse microscopy to observe if there were any changes in the levels or subcellular localisation of any of the tagged proteins in the cell pool in response to the applied treatment.
Importantly, the team found that the CRISPR-Cas9 tagging strategy they applied then enabled them to identify which proteins changed localisation by using in situ sequencing of the entire cell pool. They confirmed the known targets of these drug but also revealed unexpected changes. Particularly for perturbations of BRD4 signalling, they were able to report changes in localisations of six proteins that had previously not being recognised by any other high-throughput methods. Finally, they also showed that the method reveals expected and novel protein localisation changes as response to treatment with the approved cancer drug methotrexate.
“Our study describes a technology which, not only, for the first time, applies intron tagging to a gene pool, but is also significantly optimized in all three steps – intron tagging, cellular imaging and in situ sequencing – to enable the process in the most effective way. This method applied to chemical libraries and candidate molecules is particularly powerful in order to develop and deeply characterise drugs including the induction and inhibition of protein-protein interactions and chemical degradation. The described strategy will potentially accelerate drug discovery and have great impact on the study of global and subcellular proteome dynamics,” Kubicek explained.
The findings have been published in Genome Research.
Related topics
CRISPR, Imaging, Microscopy, Protein, Protein Expression, Proteomics
Related organisations
CeMM
Related people
Andreas Reicher, Anna Koren, Stefan Kubicek


