SARS-CoV-2 mechanism of infection revealed using cryo-electron microscopy
Posted: 8 October 2020 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have found that the surface of SARS-CoV-2 can take on at least 10 different structural states when in contact with ACE2.

The Spike (S) protein on the surface of the SARS-CoV-2 coronavirus can adopt at least 10 distinct structural states, when in contact with the human virus receptor angiotensin-converting enzyme 2 (ACE2), according to new research. The study was conducted at the Francis Crick Institute, UK.
The researchers say this new insight into the mechanism of infection will equip research groups with the understanding needed to inform studies into vaccines and treatments.
The surface of SARS-CoV-2, the virus that causes COVID-19, is covered in proteins called Spikes, which enable the virus to infect human cells. The infection begins when an S protein binds with ACE2 cell surface receptors and at later stages, catalyses the release of the virus genome into the cell. However, the exact nature of the ACE2 binding to the SARS-CoV-2 S protein remains unknown.
Now, the team has incubated a mixture of the S protein and ACE2 before trapping different forms of the protein by rapid freezing in liquid ethane. They examined these samples using cryo-electron microscopy, obtaining tens of thousands of high-resolution images of the different binding stages.
They observed that the S protein exists as a mixture of closed and open structures. Following ACE2 binding at a single open site, the S protein becomes more open, leading to a series of favourable conformational changes, priming it for additional binding. Once the S is bound to ACE2 at all three of its binding sites, its central core becomes exposed, which may help the virus to fuse to the cell membrane, permitting infection.
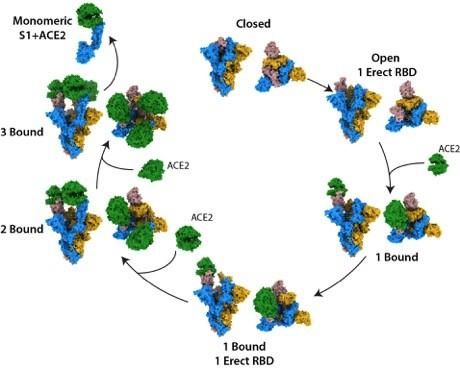

The structural states of the SARS-CoV-2 spike protein, binding to the human cell receptor ACE2 [credit: The Francis Crick Institute].
“By examining the binding event in its entirety, we have been able to characterise S protein structures that are unique to SARS-CoV-2,” said Donald Benton, co-lead author and postdoctoral training fellow at the Crick. “We can see that as the S protein becomes more open, the stability of the protein will reduce, which may increase the ability of the protein to carry out membrane fusion, allowing infection.”
The researchers hope that the more can be uncovered about how SARS-CoV-2 differs from other coronaviruses, the more targeted the development of new treatments and vaccines can be.
Antoni Wrobel, co-lead author and postdoctoral training fellow at the Crick, said: “As we unravel the mechanism of the earliest stages of infection, we could expose new targets for treatments or understand which currently available antiviral treatments are more likely to work.”
The team are continuing to examine the structures of spikes of SARS-CoV-2 and related coronaviruses in other species to better understand the mechanisms of viral infection and evolution.
The findings were published in Nature.
Related topics
Disease research, Drug Targets, Microscopy, Protein, Proteomics, Research & Development, Structural biology, Targets
Related conditions
Covid-19
Related organisations
Francis Crick Institute
Related people
Antoni Wrobel, Donald Benton



