Scientists demonstrate how COVID-19 infects human cells
Posted: 5 March 2020 | Hannah Balfour (Drug Target Review) | 69 comments
Researchers have used cryogenic electron microscopy to show that coronaviruses enter human cells through an interaction with angiotensin-converting enzyme 2 (ACE2).

Scientists exploring how coronaviruses like COVID-19 infect human cells have shown that the SARS-CoV-2 spike (S) glycoprotein binds to the cell membrane protein angiotensin-converting enzyme 2 (ACE2) to enter human cells.
COVID-19 has been shown to bind to ACE2 via the S protein on its surface. During infection, the S protein is cleaved into subunits, S1 and S2. S1 contains the receptor binding domain (RBD) which allows coronaviruses to directly bind to the peptidase domain (PD) of ACE2. S2 then likely plays a role in membrane fusion.
Chinese researchers have now used cryogenic electron microscopy (cryo-EM) to study the structure of the ACE2 when it is bound to one of its typical ligands, the amino acid transporter B0AT1 and also how the COVID-19 RBD may bind to the ACE2-B0AT1 complex. These structures have previously not been identified and could aid in producing antivirals or a vaccine that can block coronavirus infection by targeting ACE2.
The paper, published in Science, suggests ACE2 needs to dimerise to be active. The resultant homodimer has two PDs, able to bind two COVID-19 S protein trimers simultaneously.
A previous study found COVID-19 S proteins form trimers with two of the RBDs facing one direction (down) and the other facing the opposite way (up).
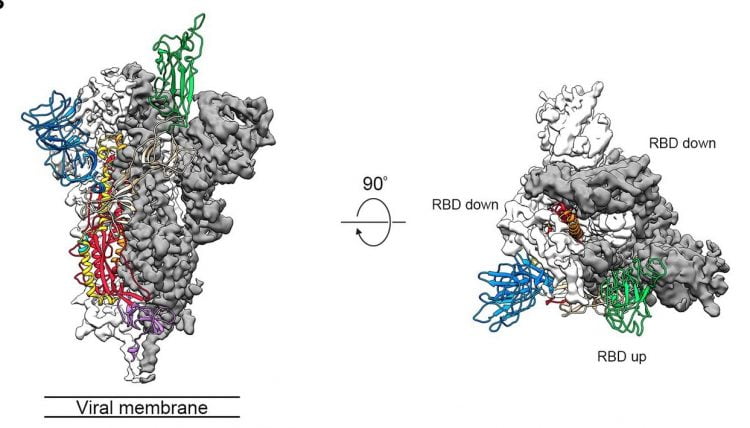

Side and top views of the pre-fusion structure of the COVID-19 S protein with a single RBD in the up conformation. The two RBD-down protomers are shown as cryo-EM density in either white or grey and the RBD-up protomer is shown in ribbons coloured green (credit: adapted from Wrapp, D, et al.).
In the current study, the team identified that the structures could only bind if the PD interacts with the up RBD.
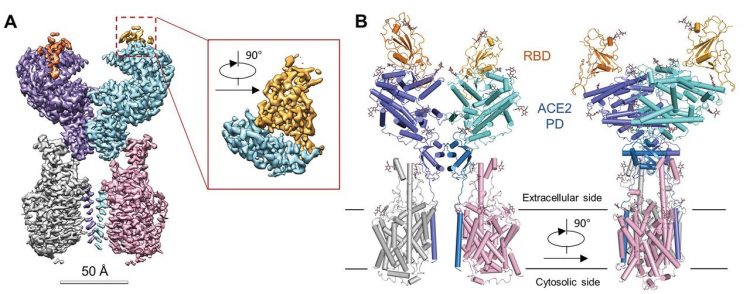

The overall structure of the RBD-ACE2-B0AT1 complex. (A) Cryo-EM map of the RBD-ACE2-B0AT1 complex. Left: Overall reconstruction of the ternary complex at 2.9 Å. Inset: focused refined map of RBD. (B) Overall structure of the RBD-ACE2-B0AT1 complex. The complex is coloured by subunits, with the protease domain (PD) and the Collectrin-like domain (CLD) coloured cyan and blue in one of the ACE2 protomers, respectively. The glycosylation moieties are shown as sticks (credit: Yan, R et al.).
They further compared how SARS-CoV-2-RBD binding is different to other SARS-CoV-RBDs binding; showing that some changes in the sequence may make associations tighter in COVID-19, while others could reduce the binding affinity.
The researchers concluded that their research could contribute to structure-based designs of decoy ligands or antibodies able to specifically target ACE2 or coronavirus spike proteins to prevent viral infection.
Related topics
Disease research, Drug Targets, Imaging, Protein, Proteomics, Research & Development, Structural biology
Related conditions
Coronavirus, Covid-19
Related organisations
Beijing Advanced Innovation Center for Structural Biology, Tsinghua University, Westlake Institute for Advanced Study, Westlake University
Related people
Renhong Yan



thats some awesomely exciting research.i love virology!!!!
We know from TCM, and indeed conventional research, that a colon-lung axis exists in the human body. I wonder, in those severely ill people requiring ventilation, if there is defective communication between the colon and the lung, which if identified and corrected, could allow the immune system to properly deal with the threat of Covid-19??
What if we could prevent the rbd’s from getting binded up by changing the shapes of the receptors ? so that the corona doesnt even recognize it ?
It is not only the shape that matters at molecular levels, but chemical signalling also, especially in the recognition context.
Can ARB agents change the number of ACE receptors in epithelial side ?
Is ACE receptor gene type on the affinity of virus on receptor ?
Ang2 receptor AT1 and ACE2 interacts to form a complex in absence of Ang2. This makes sure that all ACE2 is occupied and not available or SARS-CoV2 entry. ARBs ( .. tans)does the same thing in that it prevents Ang2 interacting with AT1 thus stabilizing AT1 -ACE2 complex , thus also preventing virus entry
Though ARBs are known to upregulate membrane expression of ACE receptors, at this point there appear no clinical significance with respect to outcomes in COVID-19 patients. See
https://www.acc.org/latest-in-cardiology/articles/2020/03/17/08/59/hfsa-acc-aha-statement-addresses-concerns-re-using-raas-antagonists-in-covid-19
Regarding Carson’s comment below the citation merely bemoans the fact that there are no experimental or clinical data demonstrating beneficial or adverse outcomes among COVID-19 patients using ACE-I or ARB medications. The article simply calls for urgent research on this issue.
Let’s try
I think that Zn ion cause such a change in the shape of ACE2. That’s why supplements with Zn are supposed to lessen the duration of cold symptoms.
Zinc has no effect on ACE 2. it inhibits the RNA polymerase enzyme.
yeah! There is no studies to show zinc has any kind immunity over this. None!
so do ACE 2 inhibitors block the same region and is there any mitigation of inflammatory response?
When ACE 2 inhibitors like in some antihypertensive medications are on board they do INCREASE the number of ACE2 sites on the cells and may have a facilitation effect to the benefit of the virus, that is why symptoms are worse when the patient with co-morbid hypertension and already are on Ace2 inhibitor. However no strict recommendation so far to discontinue Ace2 inhibitors in patients affected with COVID-19
This is from UpToDate just few days ago:
“Patients receiving ACE inhibitors/ARBs — Patients receiving angiotensin-converting enzyme (ACE) inhibitors or angiotensin receptor blockers (ARBs) should continue treatment with these agents. This approach is supported by multiple guideline panels [138-142].
There has been speculation that patients with COVID-19 who are receiving these agents may be at increased risk for adverse outcomes [143,144]. Angiotensin-converting enzyme 2 (ACE2) is a receptor for SARS-CoV-2 [145,146], and renin-angiotensin-aldosterone system inhibitors can increase ACE2 levels. Although patients with cardiovascular disease, hypertension, and diabetes may have a more severe clinical course in the setting of infection with SARS-CoV-2, there is no evidence to support an association with these agents. In addition, stopping these agents in some patients may exacerbate comorbid cardiovascular or kidney disease and lead to increased mortality [147].”
We must distinguish ACE inhibitors and ARB blockers, although they have similar effects. Don’t ARBs support COVID by revealing very active ACEs? Perhaps ACE inhibitors rather preventing infection.
hi every body what if we can replace some other things that have similar structures like human cells Ace 2 with Ace 2 that allow us bind covid 19 with that instead of human cells ace 2 ‘s .
this work’s for healing sick people but not immune them for long period of time
I was thinking of the same thing, a compound that holds the similar structure as ACE 2 receptor and has more stronger affinity.
Try to use an ACE2 selective inhibitor like MLN-4760 wich have to be effective against that virus by blockage of his key attacked receptor.
Losartan is promising and actually exhibits pretty fair efficacy in prophylactically blocking SARS-2-CoV-2 and MERS but, most patients with respiratory infections (bac-t or viral) may present in the E.R. with moderate to severe hypotension… we can stop the viral docking… but, unfortunately kill the patient.
Interesting sidebar would research Seattle area nursing homes and assess surviving patients for data on losartan (or other ACE2 receptor inhibitor) Rx’s versus angiotensin production inhibition- Lisinopril, et al)
Hypothesis would propose correlation of elderly survivors using receptor complex (blocking) competitive inhibition: Losartan dosing prior to viral exposure.
For the critical hypotensive patient an infusion
of dopamine to maintain bp could be trialed while Losartan drug is taken
can low dose of Losartan work?
https://www.ncbi.nlm.nih.gov › pub…
Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics.
by D Gurwitz · 2020
Mar 4, 2020 · Angiotensin receptor blockers as tentative SARS-CoV-2 … angiotensin receptor 1 (AT1R) blockers, such as losartan, as … that the angiotensin-converting enzyme 2 (ACE2) very likely …
(Various other workers investigated this; 2002-ish and on.)
Btw ethanol dimish B0AT1’s synthesis and in that way it can diminish the action of the virus by disturbing the ACE2-B0AT1 complex
what if they injected decoy cells that had the same structural make-up of cells that just activated when a covid attached and just engulfed it.
Lets try to change the shape of the two structures that the active site doesn’t recognize it
Losartan and other ARB’s block ACE2 receptors. Although it targets smooth endothelial cells in blood vessels mainly, it still has affinity for lung ACE2 I believe.
What is the size of the virus COVID- 19 cell. If it is 400/500 microns (?) it is very big
comparison to other Cronoviruses.
It is 125 nm in size
Worryingly ACE2 inhibitors (blood pressure medicines) may increase morbidity: https://www.thelancet.com/pdfs/journals/lanres/PIIS2213-2600(20)30116-8.pdf
Is COVID 19 extracellular initially until it begins to infect the cells of the lung membrane when it then becomes intercellular.
S1 of CONVID-2019 may be synthesized artificially and used to block the ACE2 to previde further infaction when the early human infaction happened. As the molecular size is so small that it would be easy to reach to the target cells.
I was telling it to my friend last night, about the substitute of S1 subunit and to make a compound that holds more affinity towards it than the ACE 2 receptor.
Hi,
It is posible that an angiotensine II receptor blocker/antagonist act as inhibitor to prevent coupling of covid S proteine the ACE II enzyme?
Angiotensine II receptor may over-express/activate more ACE2 on surface, leading to more complications.
Would people taking ACE inhibitors for blood pressure have any less or greater chance of infection?
Is there any aromatic compounds that have binding affininity toward RBD of virus?
Those compounds can neutralize virus
can there be a correlation between blood pressure and covid19-ace2 binding ? can diuretics decrease the bindings ?
What if we can breaks its protein present in rna so its functions turn over but how its big question….
A very big challenge is ahead for Virology / Immunology scientists and to help pharmacology to develop vaccine as well treatment medicine against this deadly COVID-19.
In silico screening (using homology modeling of RBD domain of spike protein followed by molecular docking) of edible volatile aromatic compounds library is a faster method to select the compounds that show binding affinity towards covid-9. These compounds can bind and neutralize the virus. Since they’re natural edible compounds, no need for further clinical trials, consumption also easy via inhalation.
Nitrofurantoin, an antibiotic that is used to treat urinary tract infections, has a chemical structure that can bind on the Coronavirus S-protein / ACE2 receptor and can potentially disrupt host/virus interface
So what aromatic compounds are you talking about?
we need to study metformin as a carrier substance with ACE inhibiters.
Cell regulation. Sadly overlooked.
Among the alkaloids present in the noni plant already identified, Heinicke (1985), found considerable amounts of the xeronine precursor, the proxeronine. According to the author, xeronine acts in the transformation of the molecular structure of proteins, thus presenting a wide range of functions and biological activities, since in a situation where a protein, enzyme, or a cellular signal receptor is not in its proper configuration, xeronine will promote the rearrangement of the molecular structure to its active conformation. Moreover, according to Sang et al. (2002), the presence of xeronine allows NFJ to have a number of
what if we can neutralize the surface receptors which are responsible for binding with human cell?
somehow we can inactivate the enzyme production?
Why don’t use Efavirenz o Riluzole that are a lot of electronegative
People who recovered might possibly have developed antibody against COVID-19, couldn’t their plasma be extracted to make an antidote or vaccine out of it?
i think a drug that bind to an s protein and prevent it from binding to receptor will prevent the attachment and entry of virus
Maybe there is a protein that can counteract with ACE2. A protein that counteracts with ACE2 can prevent ACE2 from actually allowing the spike cells from the coronavirus from entering ACE2. If we can find a protein that counteracts with ACE2, the spike cells from corona will have absolutely nothing to infect, because the spike cells cannot enter/infect human cells.
It is great that we’ve already found a source of how the coronavirus enters the body-through ACE2. Now I think the answer is finding another protein that can make ACE2 die out when it recognizes the spike proteins entering the cell. Making drugs that can bind to certain proteins can be dangerous, and can have long term effects to a person’s body. This is why I think finding, or even making a protein that makes ACE2 die out when it recognizes spike proteins would be a better answer. This can probably be made an injection.
In the UK, most GPs have real time computer systems from which data is already extracted by NHSE. It should be possible to quite quickly select 1000 Covid19 patients on ARBs and compare with 1000 infected controls to see whether ARBs are protective or not either in terms of infection or degree of harm. Even if the risk of hypotension is too great in extremis, it might be valuable for those nursing at home to prevent deterioration.
Hello, the idea regarding a compound that holds the similar structure as ACE 2 receptor and has more stronger affinity sound very promising.. Also, did we think why the COVID-19 became so infectious.
According to some data the virus should contain the set genes which could be part of ANY friendly bacteria or human body as important for a cell existence. This makes it for immune system hard to ID it as it looks ‘friendly’ and immune system reacts too late. Knowing the genetic build of this virus can we Identify some of those genes and possibly modify those with a compound to help the immune system ID this virus early and so help immune system fights the virus EARLY on its own vs LATE when lungs already damaged ?
How Arbidol ( Umifenovir) is relevant?
Hello, the idea regarding a compound that holds the similar structure as ACE 2 receptor and has more stronger affinity sound very promising.. Also, did we think why the COVID-19 became so infectious.
According to some data the virus should contain the set genes which could be part of ANY friendly bacteria or human body as important for a cell existence. This makes it for immune system hard to ID it as it looks ‘friendly’ and immune system reacts too late. Knowing the genetic build of this virus can we Identify some of those genes and possibly modify those with a compound to help the immune system ID this virus early and so help immune system fights the virus EARLY on its own vs LATE when lungs already damaged ?
There is decent research showing a correlation between high concentrations of ACE2 and protection against other coronaviruses (SARS). Zinc does not affect ACE but interferes with RNA viral attachment and replication and clinical trials have shown it decreases duration, frequency, and severity of rhinovirus; decreases cytokine expression; and potentiates efficacy of conventional antibiotics against pneumonia resulting in reduced mortality. Are there any virologists here willing to communicate directly with me? herbal@got.net.
Apart from Arbidol(Unifenovir), how TMPRSS 2 Inhibitor can be relevant to target at basic level?
Hello,
It is known that epitelial cell which infected by COVID 19 have a life span of approximally 7 days. Let’s assuming all cells get born at different time then if we start from today then in next 14 days all currently exising epitelial cells in lungs will die and be replaced with new ones. This likely explain why COVID 19 incubation perion is about 14 days. The virus need to develop and spread withing this 14 days window. If a person get a virus today the virus have to survive on currently living cells within next 14 days and if it does not then the virus mostly die with cells . Can we introduce some hormone or similar which would dictate Epitelial cells of lungs to divide and die faster, let’s say every 2-3 days for period of treatment ? This would give a COVID 19 only 4 to 6 days to spread and can drastically decrease of chance of the full blown infection in a patients and works as treatment for COVID 19. Also, the avarage onset of COVID 19 symptoms is 5-6 days and based on above we can suggest that if the ALL Epitelial cell life cycle in lungs will be 4 -6 days ( 2 x 2 = 4) than for this avarage type of infection it should stop it as the virus will die with dying Epitelial cells.
Does anyone know how long it takes after a virus gets into the cell, and when the first new viruses come out to infect new cells?
Also do we know how long after exposure to a virus does the individual become contagious? Is it a day, or two days, or is it 2 or 3 days before showing symptoms. Also are there any cases that the virus gets into out cells and we don’t become sick at all?
Btw, will it not damage lung early and virus to infect other healthy cells even on early death of Epi.cells of lungs?
Btw, For how much days virus can remain intact extracellular and extracellular once entered in body?
IF IF Zinc inhibits RNA Polymerase it could arrest covid replication until the cell dies naturally – as you suggest – within 7 days. Hydroxy Quinone is a zinc ionophre too. See this: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1001176
Also note that this article explains similar and the discussion mentions Zinc promoting apoptosis (cell death). https://dadamo.com/dangerous/2020/04/07/covid-19-chloroquine-zinc-and-quercetin/#comment-899
This demonstration excellent but it is not final i.e it needs further investigation .
Btw, will it not damage lung early and virus to infect other healthy cells even on early death of Epi.cells of lungs?
Btw, For how much days virus can remain intact extracellular and extracellular once entered in body?
Sorry, I meant, For how much days virus can remain intact extracellular and intracellular once entered in body?
Covid 19 is known to damage only epithelial cell which have ACE2 receptor. It rarely reported it does some damage to other cells (like brain cell, etc.)
It will definitely damage lung epithelial cell within 14 days. The point here was that if we could reduce Artificially the lifespan of the cell so virus does not have time to spread. Could we possibly use HGH [ormone] , etc ?
If the virus could live longer than 14 days Inactive somewhere we should see symptoms of Covid 19 showing up after 14 days and it rarely happens (some rare reports show that it could be 23 days for symptoms to show up)
Sorry, I meant, For how much days virus can remain intact extracellular and intracellular once entered in body?
Elderberry? Is it true that a compound in Elderberry binds to the spike proteins and destroys them, rendering the virus incapable of piercing a cell?
https://www.healthline.com/nutrition/elderberry#health-benefits
Aldosterone
a corticosteroid hormone which stimulates absorption of sodium by the kidneys and so regulates water and salt balance.
Angiotensin
a protein whose presence in the blood promotes aldosterone secretion and tends to raise blood pressure.
Glycoprotein
any of a class of proteins that have carbohydrate groups attached to the polypeptide chain. Also called glycopeptide
*Invasive Method* In Covid 19 glycoproteins bind to the cell membrane protein angiotensin-converting enzyme 2 (ACE2) to enter human cells. *Glycoprotein inhibitor… possible preventive method*.
COVID-19 has been shown to bind to ACE2 via the S protein on its surface. During infection, the S protein is cleaved into subunits, S1 and S2. S1 contains the receptor binding domain (RBD) which allows coronaviruses to directly bind to the peptidase domain (PD) of ACE2. S2 then likely plays a role in membrane fusion.
ACE2
is an enzyme attached to the outer surface (cell membranes) of cells in the lungs, arteries, heart, kidney, and intestines. ACE2 lowers blood pressure by catalysing the hydrolysis of angiotensin II (a vasoconstrictor peptide) into angiotensin. ACE2 counters the activity of the related by reducing the amount of angiotensin-II and increasing Ang(1-7) making it a promising drug target for treating cardiovascular diseases.
Hydrolysis
the chemical breakdown of a compound due to reaction with water
Interferon (IFNs)
IFNs belong to the large class of proteins known as cytokines, molecules used for communication between cells to trigger the protective defenses of the immune system that help eradicate pathogens. Interferons are named for their ability to “interfere” with viral replication by protecting cells from virus infections.
If we can speed up the immune response through use of interferon response therapy while inhibiting the COVID 19 virus by using a glycoprotein IIb/IIIa inhibitors (would this allow the immune system to establish enough antibodies to develop in order to provide future immunity?)
Which macro-molecules are affected in the human cells that COVID-19 infect?
How many electron contain a virus?