Researchers work to create a COVID-19 viral epitope map
Posted: 13 March 2020 | Hannah Balfour (Drug Target Review) | No comments yet
The team used data from SARS-CoV to identify possible viral epitopes that vaccines could include to stimulate an immune response.

Immunologists have used data from the SARS-CoV coronavirus, the closest relation to SARS-CoV-2 – currently causing the COVID-19 outbreak, to identify regions of the virus that the immune system’s B and T cells could react to. The researchers say their discoveries provide key information for vaccine design.
Vaccine strategies that specifically target these regions could generate immunity… that’s relatively resistant to ongoing virus evolution”
The paper published in Cell, Host and Microbe provides the first analysis of potential immune system targets that need to be incorporated into vaccines. The researchers from La Jolla Institute (LJI) for Immunology and the J. Craig Venter Institute, both US, used data from the well characterised SARS-CoV coronavirus to suggest which viral epitopes (small molecular features on the viral particle) the cells of the immune system may allow recognition and activation.
“Right now, we have limited information about which pieces of the virus elicit a solid human response,” says the study’s lead author Dr Alessandro Sette, a professor in the Center for Infectious Disease and Vaccine Research at LJI. “Knowing the immunogenicity of certain viral regions, or in other words, which parts of the virus the immune system reacts to and how strongly, is of immediate relevance for the design of promising vaccine candidates and their evaluation.”
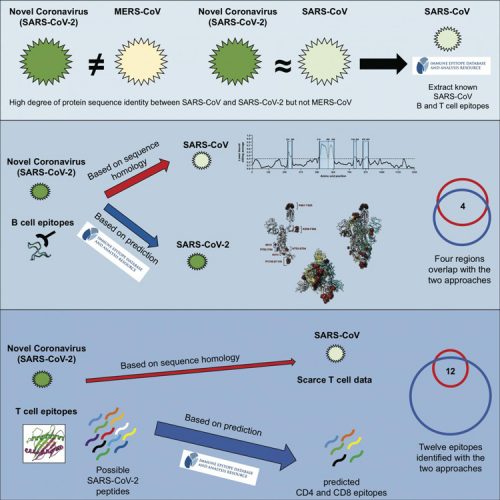

Existing data from known coronaviruses can be used to predict which parts of SARS-CoV-2 are capable of activating the human immune system (credit: Grifoni et al./Cell Host & Microbe).
These epitopes also enable the formation of memory cells which drive a faster response if the same structures are encountered again. A map of these viral epitopes and their relative immunogenicity could benefit researchers attempting to design or modify vaccines to protect against COVID-19.
In their study, the team used available data from the LJI-based Immune Epitope Database (IEDB), which contains over 600,000 known epitopes from some 3,600 different species, and the Virus Pathogen Resource (ViPR), a complementary repository of information about pathogenic viruses. The team compiled known epitopes from SARS-CoV and mapped the corresponding regions to SARS-CoV-2.
“We were able to map back 10 B cell epitopes to the new coronavirus and because of the overall high sequence similarity between SARS-CoV and SARS-CoV-2, there is a high likelihood that the same regions that are immunodominant in SARS-CoV are also dominant in SARS-CoV-2 is,” explained the study’s first author Dr Alba Grifoni, a postdoctoral researcher in the Sette lab.
The B cell epitopes identified were:
- Five on the spike glycoprotein – the crown-shaped projection from the surface of the virus
- Two in the membrane protein – the membrane protein is embedded in the membrane enveloping the shell of the virus
- Three in the nucleoprotein – the shell of the virus is formed of nucleoproteins.
In a similar analysis, T-cell epitopes were also mostly associated with the spike glycoprotein and nucleoprotein.

To substantiate the SARS-CoV-2 T cell epitopes identified based on their homology to SARS-CoV, Dr Grifoni compared them with epitopes pinpointed by the IEDB. Using this approach, she was able verify 12 out of 17 SARS-CoV-2 T cell epitopes identified based on sequence similarities to SARS-CoV.
“The fact that we found that many B- and T-cell epitopes are highly conserved between SARS-CoV and SARS-CoV-2 provides a great starting point for vaccine development,” said Dr Sette. “Vaccine strategies that specifically target these regions could generate immunity that’s not only cross-protective but also relatively resistant to ongoing virus evolution.”
Related topics
Analysis, Disease research, Drug Targets, Immunology, Protein, Protein Expression, Proteomics, t-cells, Vaccine
Related conditions
Coronavirus, Covid-19, Severe Acute Respiratory Syndrome (SARS)
Related organisations
J. Craig Venter Institute, La Jolla Institute (LJI) for Immunology
Related people
Dr Alba Grifoni, Dr Alessandro Sette


