A2aAR protein holds clues for better drug design
Posted: 2 January 2018 | Dr Zara Kassam (Drug Target Review) | No comments yet
Scientists have investigated a key protein used in drug design and discovered dynamic structural features that may lead to new ways to target diseases…

Scientists at The Scripps Research Institute (TSRI) have peered deep into the heart of a key protein used in drug design and discovered dynamic structural features that may lead to new ways to target diseases. The protein, called the A2A adenosine receptor (A2aAR), is a member of the G-protein-coupled receptor (GPCR) family, which are the targets of roughly 40 percent of all approved pharmaceuticals.
The new, more detailed image of A2aAR’s signalling mechanism reveals key parts of its inner workings, including an amino acid that acts like a “toggle switch” to control signals across the cell membrane.
“This basic knowledge is potentially helpful for improving drug design,” says Nobel laureate Kurt Wthrich, the Cecil H. and Ida M. Green Professor of Structural Biology at TSRI and senior author of the study.
Imaging technique reveals how protein changes shape
All human cells contain A2aAR and other GPCRs embedded in their plasma membrane. More than 800 GPCRs have been discovered in the human body, and each has a role in regulating a bodily function. For example, A2aAR regulates blood flow and inflammation and mediates the effects of caffeine. A2aAR is also a validated target for treating Parkinson’s disease and a relatively new target for targeting cancers.
“GPCRs do just about everything you can imagine,” says Dr Wthrich. “But for a long time, drug design was being done without knowing how GPCRs looked.”
For the new study, the researchers aimed to better understand the relationship between A2aAR function and dynamic changes in its structure to help inform drug design.
The research built on previous studies where scientists used an imaging technique called x-ray crystallography to determine A2aAR’s three-dimensional structure. The images showed that A2aAR looks like a chain that crisscrosses the cell membrane and has an opening on the side facing out of the cell. The region of the GPCR structure that sticks out of the membrane interacts with drugs and other molecules to signal to partner proteins inside the cell.
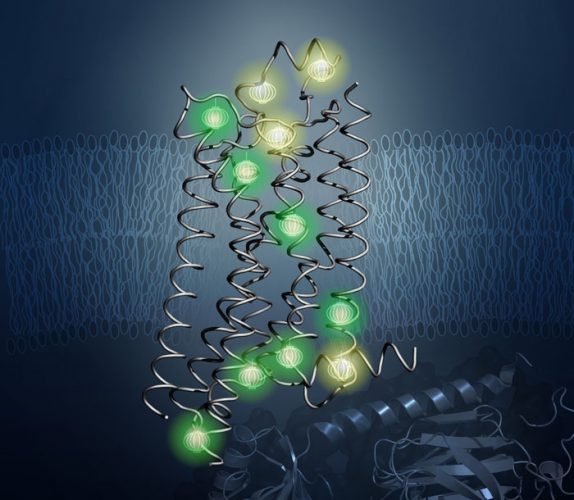

Probes (shown glowing here) revealed the inner architecture of the protein A2aAR in the new study. CREDIT: Kurt Wuthrich and Matthew Eddy, The Scripps Research Institute
Although crystal structures provided a key outline of the receptor’s shape in inactive and active-like states, they could not show motion and changes in structure when A2aAR meets new binding partners, such as pharmaceutical candidates. In short, the researchers in the new study needed to investigate why A2aAR works the way it does.
To solve this problem, the researchers used a technique called nuclear magnetic resonance (NMR) spectroscopy, which creates strong magnetic fields to locate the positions of probes in a sample. With NMR, scientists can determine the structures of proteins and study their dynamic properties in solution at physiological temperatures–the way they exist in the human body.
Dr Matthew Eddy, the first author of the study, used NMR to observe A2aAR in many different conformations, shedding light on how it changes shape on the surface of human cells in response to drug treatments.
Importantly, NMR let the team visualise changes in the internal architecture of A2aAR. This took them beyond previous solution NMR studies, which focused on the technically less demanding observation of NMR-observable probes attached to flexible parts of GPCRs, mostly located at or near the surface of the receptor. The approach in the new study enabled researchers to follow the effects of drug binding at the extracellular surface on changes in protein structure and dynamics at the intracellular surface–the structural basis of signal transfer–across the heart of the GPCR.
It was like the researchers had seen a car, and with NMR, they could finally inspect its engine.
Rethinking how we design drugs
Two details in A2aAR’s structure gave researchers insight into how future drugs could manipulate the receptor. One key finding was that replacing one particular amino acid in the receptor’s centre destroyed the receptor’s ability to send signals into the cell.
“With this finding, we can say ‘A-ha! It is this change in structure that kills the signalling activity.’ Maybe we can make a change in a drug to overcome this limit,” says Wthrich.
The researchers also revealed the activity of a “toggle switch” in A2aAR. Previous studies suggested that one of the tryptophan amino acids in A2aAR flips up and down in concert with A2aAR’s activity. With NMR, the scientists directly observed this unique tryptophan as it changed orientations in response to different drugs. Chemists could potentially modify drugs to manipulate this switch and control A2aAR signalling.
The researchers emphasise that this new study is potentially relevant for much of the large family of GPCRs. In fact, structural details from this study could apply to more than 600 “class A” GPCRs in our bodies.
The findings were published in the journal Cell.
Related topics
Disease research, Drug Discovery, Drug Discovery Processes, Drug Targets, Imaging, Nuclear Magnetic Resonance (NMR), X-ray crystallography
Related organisations
The Scripps Research Institute (TSRI)
Related people
Kurt Wthrich


