VP3 protein: revealing the structure of a potential rotavirus drug target
Posted: 17 April 2020 | Hannah Balfour (Drug Target Review) | No comments yet
Researchers hope that by revealing the rotavirus VP3 protein structure and mRNA capping functions, novel antivirals could be designed to prevent or combat rotavirus infections.

After more than three decades of research, a team led by scientists at Baylor College of Medicine, US, have unveiled the structure of the rotavirus VP3 protein, a key target for potential therapeutics.
The rotavirus, and several other viruses, use enzymes to cap their messenger RNA (mRNA). This process disguises the viral mRNA as the hosts and enables it to interact with host cellular machinery to facilitate the replication of the viral genome. Capping their mRNA also enables viruses to evade the host immune system.
“Viruses cannot replicate on their own. They take over the machinery of the cells they infect to produce viral particles,” said co-corresponding author Dr B. V. Venkataram Prasad, professor and Alvin Romansky Chair of the Verna and Marrs McLean Department of Biochemistry and Molecular Biology and professor of molecular virology and microbiology at Baylor.
Drug Target Review has just announced the launch of its NEW and EXCLUSIVE report examining the evolution of AI and informatics in drug discovery and development.
In this 63 page in-depth report, experts and researchers explore the key benefits of AI and informatics processes, reveal where the challenges lie for the implementation of AI and how they see the use of these technologies streamlining workflows in the future.
Also featured are exclusive interviews with leading scientists from AstraZeneca, Auransa, PolarisQB and Chalmers University of Technology.
In the study published in Science Advances the team found that the VP3 protein – which conducts the capping of the rotavirus mRNA – consists of four molecular modules which, unlike other viruses, integrate five enzymatic activities necessary for capping mRNA. The team hope their discovery could be used to design antiviral drugs to combat rotavirus infection by limiting the capping capability of the VP3 protein.
Unveiling the structure and function of VP3
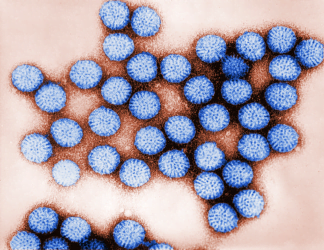

Digitally-colourised transmission electron microscopic (TEM) image of rotavirus particles [credit: CDC / Bryon Skinner].
Dr Dilip Kumar, study first author and a postdoctoral fellow in the Prasad lab, explained: “Combining cryo-electron microscopy, X-ray crystallography and biochemical assays, we discovered that VP3 has all the enzymatic activities required to effectively cap rotavirus mRNA. We also found that, opposed to what has been seen in many viruses that have individual proteins for each enzymatic activity, rotavirus integrates all the enzymatic activities as modules into one protein, VP3.”
Study co-author Dr Mary K. Estes, Cullen Foundation Endowed Chair and Distinguished Service Professor of molecular virology and microbiology at Baylor, added: “What makes rotavirus different from most other viruses is that capping takes place inside the tight confines of the viral capsid, the protein shell of the virus, that is inside the cell. Once capping is complete, viral mRNA exits the capsid through channels and enters the cytoplasm of the cell where it engages the protein-making machinery to produce viral proteins.”
Kumar continued: “We were able to isolate VP3 and its modules and show that they self-assemble into a stable molecule capable of capping at least two mRNAs simultaneously, making the process efficient.”
Moving forward
The researchers hope that this new understanding of the structure and functions of the VP3 protein will allow antiviral drugs to be designed to more effectively prevent or treat rotavirus infections.
Prasad concluded: “For nearly 30 years, my lab and Dr Estes’s have collaborated to unravel the complex structure of rotavirus and better understand how it works. Determining the structure and function of VP3 is a major milestone that opens new doors for continuing our collaboration.”
Related topics
Crystallography, Disease research, Drug Targets, Enzymes, Imaging, Microscopy, Molecular Targets, Protein, Structural biology
Related conditions
Rotavirus
Related organisations
Baylor College of Medicine
Related people
Dr B. V. Venkataram Prasad, Dr Dilip Kumar, Dr Mary K. Estes



