Researchers reveal how SARS antibody binds to COVID-19 coronavirus
Posted: 7 April 2020 | Victoria Rees (Drug Target Review) | No comments yet
The interaction between a SARS antibody called CR3022 and the COVID-19 coronavirus has been mapped by researchers, revealing a viral vulnerability.

According to researchers, an antibody recovered from a survivor of the severe acute respiratory syndrome (SARS) epidemic, named CR3022, has revealed a potential vulnerability of the COVID-19 coronavirus.
The study, conducted at Scripps Research, US, is the first to map a human antibody’s interaction with COVID-19 at near-atomic-scale resolution. Although the antibody was produced in response to an infection of SARS, which is caused by SARS-CoV, it cross-reacts with the new coronavirus, SARS-CoV-2, say the researchers.
The structural mapping revealed a nearly identical site at the Spike (S) protein on both coronaviruses to which the antibody binds, suggesting a functionally important and vulnerable site for this family of viruses.
“The knowledge of conserved sites like this can aid in structure-based design of vaccines and therapeutics against SARS-CoV-2, which would also protect against other coronaviruses – including those that may emerge in the future,” said the study’s senior author Professor Ian Wilson, Chair of the Department of Integrative Structural and Computational Biology at Scripps Research.
“Our ultimate goal here is to obtain structural information on antibodies and their binding sites and use that to guide SARS-CoV-2 vaccine design, just as our lab has done with influenza and HIV,” said the study’s co-first author Dr Nicholas Wu, a postdoctoral research associate in the Wilson lab.
A report from Chinese scientists earlier this year indicated that CR3022 cross-reacts against SARS-CoV-2. Wilson’s team used their structural mapping expertise to determine how the antibody binds to SARS-CoV-2.
A key finding is that the antibody’s binding site is highly similar between the two coronaviruses, differing by just four amino acids. That high degree of similarity implies that the site has an important function that would be lost if it mutated significantly.
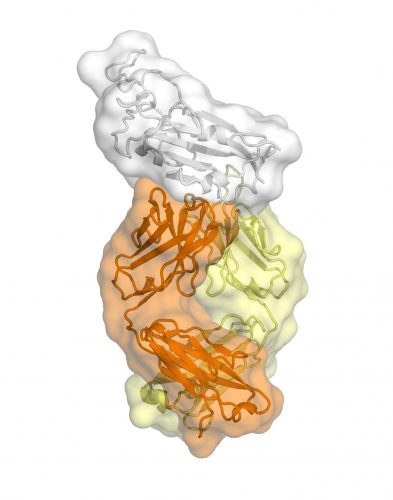

This is an antibody CR3022 bound to the receptor binding domain of SARS2-CoV-2 [credit: Meng Yuan and Nicholas Wu].
However, the site’s function remains unknown. The researchers’ analysis found that the antibody binding site is relatively remote from the part of the virus that binds to cell-surface protein receptors in preparation for penetrating cells in the lungs. That suggests that, at least for SARS-CoV, CR3002 neutralises the virus’s ability to infect cells in some indirect way.
“We found that this region is usually hidden inside the virus and only exposed when that part of the virus changes its structure, as it would in natural infection,” said co-first author Dr Meng Yuan, also a research associate in the Wilson lab.
Despite the slightness of difference between the two coronaviruses, the antibody binds much less tightly to SARS-CoV-2 than it does to the SARS virus and cannot neutralise SARS-CoV-2 in lab dish tests as it does SARS-CoV.
The findings still suggest however that the binding site for this antibody on SARS-CoV-2 is a site of vulnerability and that antibodies binding it more tightly would plausibly succeed in neutralising the virus. Such neutralising antibodies, if developed into therapies, could be used to treat COVID-19 patients and to provide temporary protection from the virus to uninfected individuals.
The fact that this binding site is highly conserved between SARS-CoV and SARS-CoV-2 also hints that there may be antibodies, still to be discovered, that can effectively neutralise both viruses, say the researchers.
The study is published in Science.
Related topics
Antibodies, Antibody Discovery, Disease research, Drug Targets, Protein, Proteomics, Research & Development, Structural biology, Targets
Related conditions
Coronavirus, Covid-19
Related organisations
Scripps Research
Related people
Dr Meng Yuan, Dr Nicholas Wu, Professor Ian Wilson



