A close-up view of the Ebola virus life cycle
Posted: 26 June 2015 | Victoria White
A new study by scientists from the Scripps Research Institute reveals a key part of the Ebola virus life cycle at a higher resolution than ever before…
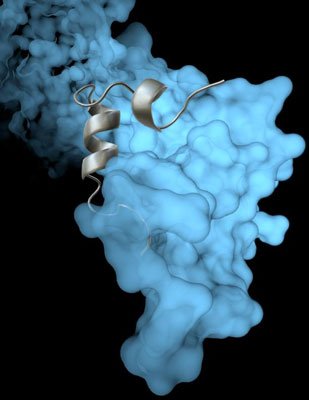

The Scripps Research Institute team’s high-resolution image revealed how a viral protein called VP35 helps protect the Ebola virus from the body’s immune system. Photo courtesy of The Scripps Research Institute.
A new study reveals a key part of the Ebola virus life cycle at a higher resolution than ever before.
The research sheds light on how Ebola virus assembles—and how researchers might stop the often-fatal infection.
“This higher resolution is critical for design of much-needed antiviral therapeutics,” said Erica Ollmann Saphire, professor at The Scripps Research Institute (TSRI) and director of the Viral Haemorrhagic Fever Immunotherapeutic Consortium. “These structures provide the blueprints that we need to see key vulnerabilities to attack.”
The new study builds on previous work in Saphire’s lab showing that a viral protein called VP35 has a role in protecting both Ebola virus and its ‘cousin’, the deadly Marburg virus, from the immune system. VP35 chaperones a viral protein, helping it coil and form a protein shell (nucleocapsid) around the virus’s genetic material. With a virus’s genetic material blocked from view, the human immune system cannot mount an effective defense.
Findings could have significance beyond the Ebola virus
Until now, scientists had not been able to see the coiling process in great detail. But using an imaging technique called x-ray crystallography, Saphire and her colleagues were able to show exactly how VP35 helps the viral protein that creates the capsid.
The new structure also reveals how the VP35 protein prevents the nucleocapsid from assembling incorrectly. The researchers could even see key atoms and structures called side chains—crucial pieces for moving forward with structure-based drug design.
The researchers believe these findings could be significant for more than just Ebola virus. “The structure we revealed is likely conserved across all the filoviruses: Marburg, Sudan, Bundibugyo, Reston and Ebola,” said Saphire.
TSRI Research Associate Robert Kirchdoerfer added that the new understanding of viral assembly could be applied to Mononegavirales, an order of viruses that includes measles and rabies.
The study findings are published in Cell Reports.
Related topics
X-ray crystallography
Related conditions
Ebola
Related organisations
The Scripps Research Institute (TSRI)



