Data, data and still more data: the challenges of imaging and analysing 3D cell models
Posted: 4 June 2020 | Jeremy Simpson (University College Dublin), Margaritha Mysior (University College Dublin) | No comments yet
The latest generation of three-dimensional (3D) cell models offer new and exciting possibilities for pharmaceutical research. However, deciding how to capture sufficient information in the most efficient way from an experiment without making the data size overwhelming can prove challenging. Margaritha Mysior and Jeremy Simpson discuss the opportunities and challenges faced by researchers currently working in this field.


The 21st century is rapidly becoming known as the century of data. The data that infiltrate our daily lives come from commerce, finance and social media; all organisations dealing with such vast amounts of data face a constant struggle to not only cope with ever increasing volumes, but also carry out meaningful analysis and interpretation. This issue is becoming progressively more apparent in research science, with large-scale datasets from sequencing, genomics, proteomics and imaging experiments. In the specific case of biological imaging, it is the emergence of the use of 3D cell models (eg, spheroids and organoids) that is the most significant in terms of the size of the datasets produced. Here, we discuss the potential that such experiments offer biomedical science research and also the data challenges faced.
Why does the imaging of 3D cell models produce such a large amount of data and why are these experiments important for our greater understanding of cellular processes? One reason is that during the last decade it has becoming increasingly clear that 3D cell models offer several advantages over traditional two-dimensional (2D) cell monolayers. Particularly noteworthy is that methodologies have been developed to create 3D cell models from a wide variety of both normal and tumour cells, they can be composed of multiple cell types within a single assembly and that the cell interactions that form more closely mimic how tissues are constructed in vivo. In addition, 3D cell models can be applied to a growing range of research areas, such as the study of fundamental processes in tissues and organs, dissecting the pathways that drive cancer and as more physiological models for pre-clinical assessment of drug delivery and toxicology. Together, such models have the potential to provide significantly more accurate information compared to cell monolayers and they can take into account the fact that the human body not only consists of different cell types but that their spatial organisation and interactions are essential for overall function. However, to fully maximise the information that can be extracted from spheroid and organoid models, imaging approaches must not only provide data from the full depth of the entire cell assembly, but also retain information from individual cells – in some cases at the subcellular and organelle scale. Inevitably, this leads to datasets that are on a magnitude far greater than anything obtained from monolayer cultures.
Classical 2D cell culture and the power of quantitative measurements

Classical 2D cell culture has served biomedical science for well over half a century, largely as a result of the pioneering work by scientists such as Wilton Earle, George Gey, Harry Eagle and Renato Dulbecco, who together developed some of the first in vitro cultured cell lines and the nutritional media required to sustain them.1 The majority of the mammalian cultures that they worked with grew in the form of monolayers, which was a convenient and cost-efficient format for the establishment of many types of assays, eventually leading to their adaptation into multi-well plates compatible with optical microscopy approaches. The reagents and wider technology around fluorescence in cell-based assays has also developed at a rapid pace in recent years, spawning the quantitative imaging technologies termed high-content screening (HCS) and high-content analysis (HCA). These techniques utilise automated microscopes to rapidly acquire large fluorescence datasets from cells, after which associated software is used to extract highly parallel quantitative descriptors of the fluorescent patterns. These quantitative measurements include the intensity (and therefore abundance) of different markers, their size, distribution, localisation and co-localisation, as well as providing a functional readout of the biological event under study. Combining appropriate parameters allows HCA to aid in the understanding of pathways and processes within the cell and enable an easy comparison of treatments or perturbations. HCA has therefore revolutionised many aspects of the pharmaceutical development pipeline, facilitating not only the high-throughput testing of lead compounds in a wide range of cell-based assays, but providing detailed quantitative multi-parametric readouts for toxicity and efficacy.
However, despite their relative ease of use, there are limitations to using cells grown in monolayers. Most importantly, they do not take account of the natural way in which tissues and organs are built or how the different cells communicate and interact with one another in a multi-dimensional space. On the other hand, 3D models can potentially overcome these critical limitations, as they can be composed of multiple cell types, replicate physiological cell-cell contacts and secrete extracellular matrix. It is therefore clear that they offer distinct advantages in terms of being a more accurate model for pre-clinical prediction of therapeutic efficacy and toxicity.2 While the biological case for their use is compelling, from an imaging and image analysis perspective they present new challenges.
Challenges in acquisition and analysis of 3D image data


The current generation of automated HCS microscopes now offer the realistic potential of image acquisition from 3D cell assemblies on a large scale. Such instruments also provide the possibility of imaging using high resolution objectives with the capacity to capture whole spheroid or organoid information as well as subcellular information from individual cells. Therefore, the major challenge currently is how to most efficiently capture sufficient information from the experiment without producing an overwhelming data size. Careful consideration needs to be given to whether imaging of the entire depth of the spheroid or organoid is required or whether sampling a selection of optical slices through the structure will provide adequate information.3 Similarly, a decision has to be made with respect to the area of the well to be imaged; specifically, whether information is needed from the entire well or only a portion of it. The impact of this is illustrated in Figure 1, comparing the data sizes for a three-colour cell‑based assay carried out in monolayer-grown cells (2D), versus in spheroids (3D). For a single well of data in a 96‑well plate, considering the entire well (70 fields-of-view), moving from a 2D to 3D assay dramatically increases the data size from 969MB to 29GB. When this is scaled to a modest screen of 300 96-well plates, the difference in data volume becomes truly immense, namely 28TB versus 840TB. Sampling a portion of the well by acquiring fewer fields-of-view or decreasing the number of optical slices collected reduces the data volume, although the overall data size remains challenging. Consideration also needs to be given to the storage and later retrieval of the datasets. While hard disk drive capacity is increasing exponentially year-on-year, it may be unfeasible to continue to store primary image data from large screens for extended periods of time.
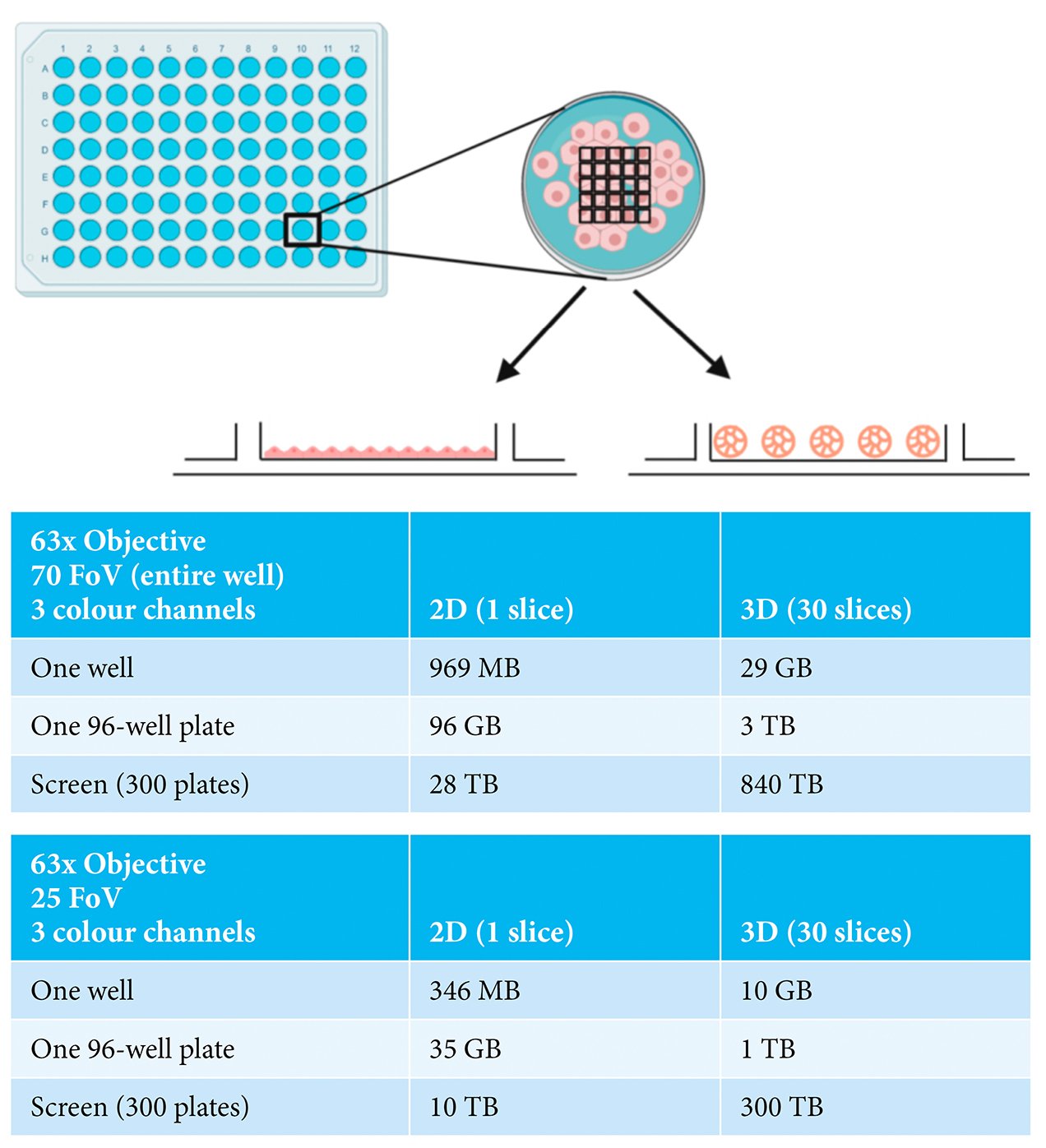

Figure 1: Examples of imaging experiments carried out in either 2D- or 3D-grown cells showing
indicative data size ranges depending on numbers of fields-of-view (FoV), numbers of
wells and numbers of plates. Made with BioRender.com.
Once the image acquisition parameters have been finalised, the next issue to address relates to how the data will be analysed. Unlike the scenario in monolayer cells, 3D models provide the possibility of investigating cellular processes in multiple dimensions. Although the volumetric component of each cell and spheroid provide important information, this does not come without its challenges. In the first instance, accurate volumetric analysis needs sufficient optical planes to have been acquired and their subsequent processing requires major computing power. In addition, HCA relies on software to correctly identify the individual cells through a process termed segmentation, followed by the correct assignment of each object (such as an organelle or structure) to each cell. These segmentation and assignment algorithms are well-established and highly robust for monolayer cells, but are comparatively still in their infancy for use with spheroids or organoids (Figure 2) – not to mention that they require significantly greater computer processing capacity when applied in 3D. Poor segmentation or object assignment would result in all downstream measurements being incorrect, potentially leading to false interpretations from the experiment. However, once such challenges are overcome, the power of these cell models can be fully exploited. For example, they can be used to understand the mechanisms and efficacy of how a pharmaceutical product passes through the multiple cell layers of a spheroid or how it induces varying levels of toxicity to the different types of cells in an organoid. When the imaging is carried out with sufficient resolution, quantitative volumetric analysis of individual organelles or subcellular structures within each cell across the entire spheroid or organoid becomes possible, potentially providing mechanistic insights that are simply not possible to be discovered in monolayer-grown cells. What is also becoming clear is that drug responses can be very different between cells grown in 2D versus 3D. Therefore, the more physiological the in vitro models are at pre-clinical stages, then the greater the chance of eliminating the drug before it reaches animal or clinical trials.
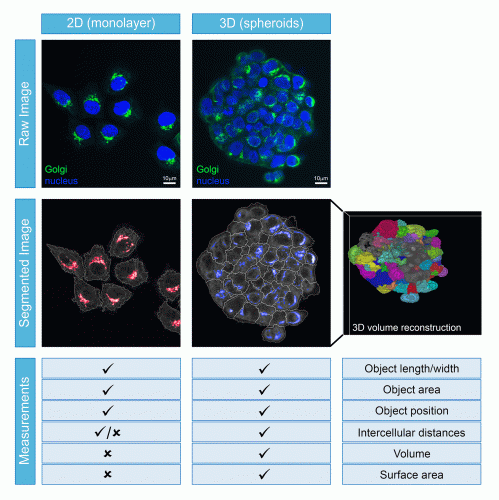

Figure 2: Comparison of cell segmentation ability and measurements that can be obtained from cells growing as monolayers or spheroids.
The latest generation of 3D cell models offer new and exciting possibilities for pharmaceutical research. The combination of innovative cell culture and imaging technologies together provide the opportunity for much improved pre-clinical assessment of pharmaceutical performance, although certain hurdles remain. Not least of these is that the data generated from imaging and subsequent analysis is potentially of a scale never seen before in biological experiments. Large-scale screening in spheroids or organoids is now moving into Terabyte and soon Petabyte territory, presenting data handling, management and storage challenges that need to be overcome. Solutions to these problems need to be found quickly, so that we can truly realise the potential of this new generation of cell models.
About the authors


Margaritha M Mysior graduated with a BSc (2015) and MSc degree (2017) in Molecular Biology from the University of Applied Sciences Gelsenkirchen, Bocholt and Recklinghausen, Germany. She spent the final year of her MSc degree in the lab of Jeremy Simpson at University College Dublin, Ireland, characterising several newly identified regulatory proteins of the Golgi-ER retrograde pathway in mammalian cells. She started her PhD in the same lab in 2017 and is currently developing a number of imaging-based assays to study the endomembrane system in a 3D cell model.


Jeremy C Simpson carried out his PhD work at the University of Warwick, UK, and post-doctoral work at the Scripps Research Institute in San Diego, USA, the ICRF in London, UK, and the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany. In 2008 he was appointed as Professor of Cell Biology at University College Dublin, Ireland. His lab currently applies high-throughput imaging technologies to study membrane trafficking processes and how cells interact with nanomaterials. He has authored over 100 peerreviewed articles, runs the UCD Cell Screening Laboratory (www.ucd. ie/hcs) and is a principal investigator in the CÚRAM Centre for Research in Medical Devices (www.curamdevices.ie).
References
1. Yao T, Asayama Y. Animal-cell culture media: History, characteristics, and current issues. Reproductive Medicine and Biology. 2017;16(2):99-117.
2. Shen J, Youhanna S, Zandi Shafagh R, Kele J, Lauschke V. Organotypic and Microphysiological Models of Liver, Gut, and Kidney for Studies of Drug Metabolism, Pharmacokinetics, and Toxicity. Chemical Research in Toxicology. 2019;33(1):38-60.
3. Cutrona M, Simpson J. A High‐Throughput Automated Confocal Microscopy Platform for Quantitative Phenotyping of Nanoparticle Uptake and Transport in Spheroids. Small. 2019;15(37):1902033.
Related topics
Analysis, Analytical techniques, Big Data, Cell-based assays, High-content assays, Imaging, Organoids, Technology
Related organisations
Molecular Devices
Related people
George Gey, Harry Eagle, Renato Dulbecco, Wilton Earle


