Computational methods for drug repositioning
Posted: 17 February 2016 | Anaum Rashid (University of Sunderland), Hannah Smith (University of Sunderland), Ken McGarry (University of Sunderland) | No comments yet
Bioinformatics and computational techniques are now experiencing increased attention as a means of improving the costly and time-consuming drug development process…


Recently, drug repositioning has been suggested as an interim way of harnessing existing drugs for different diseases or medical conditions than the original target. The numerous online databases of chemicals, protein interactions, side effects, drug mechanisms and drug toxicology information make drug repurposing a suitable application area for computational intelligence. Thus, in-silico analysis can be used as a useful first stage to screen potential candidate drugs for possible redeployment. We hypothesise that the side effects of the current Alzheimer’s drugs can be used to screen the SIDER (side effects) database for potential drug candidates with similar side effects; that is to say they are probably affecting the same protein targets. Thus, a previously harmful situation has been turned into a mechanism for drug repositioning.
Our system uses complex graphical network techniques to analyse drugs with known side effects and compares the proteins involved in these side effects with proteins known to be identified with other diseases. Our intention is to find potential candidates for treating Alzheimer’s disease.
Databases and online repositories
Researchers have examined the utility of considering drug side effects as way of repositioning with varying degrees of success1,3. Our method differs in that we also consider the protein interactions and other possible side effects. Our initial research focused building a list of side effects experienced by patients receiving the three drugs donepezil, galantamine and rivastigmine, commonly used to treat Alzheimer’s disease. These were used to establish the common side effects, which were then used to identify the drug candidates. Our systems data flow and data integration are shown in Figure 1.
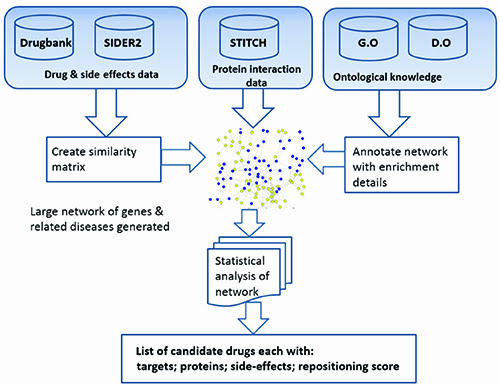

Figure 1: System overview and data integration aspects.
We used a number of databases but the most important was the SIDER2 database; this important repository contains hundreds of drugs and thousands of their known side effects on humans9. The latest release contains 4,192 side effects and 996 drugs, information that was extracted from public documents and package inserts. The database is structured into drug classes and types of side effect and has been very useful in drug target prediction. The side effects pertaining to the three Alzheimer’s drugs were obtained from this database. In Figure 2, a Venn diagram illustrates the common side effects shared by the three drugs and those which are unique to each drug. The decision was made to use only the side effects common to all three drugs which meant that 97 unique side effects were available for study.
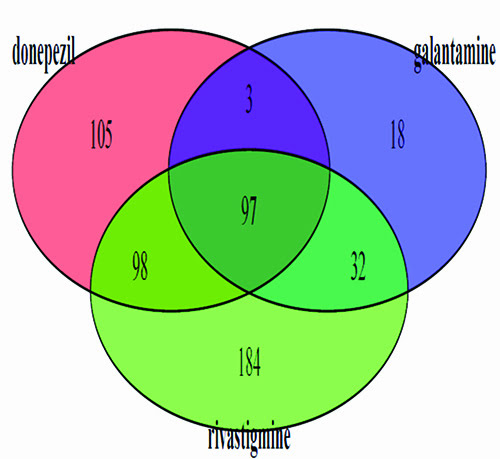

Figure 2: Venn diagram indicating the joint side effects of three Alzheimer’s drugs. The 97 side effects common to all three drugs (centre of diagram) were used as a list to search the SIDER2 database.
The DrugBank database was also utilised because it lists the majority of drugs that are prescribed or have reached the clinical trials stage and is widely used by those developing drugs, chemists, pharmacologists and those involved in pharmaceutics research10. Each drug is listed with its main targets, known off-targets, the chemical structure and other properties. Gene ontology (GO) and disease ontology (DO) are used to annotate the proteins and drug targets with additional information useful for a deeper interpretation of the biological processes and structures4,5. The DO database gives insight into 8,043 inherited, developmental and acquired human diseases6,7.
To gain a better understanding of biological plausibility, our system searched the GO to a depth of three levels, using the biological process (BP) section of GO database. As seen in Table 1, the results for the donepezil group of 16 proteins are displayed. Only the first 10 are shown, in descending order of importance. For example, for the first entry, GO:0044763, which corresponds to single-organism cellular process, all 16 proteins are known to cooperate in this activity, while the last entry in the table refers to response to chemical with 13 of the 16 proteins participating in this function. In total there were 121 identified activities in BP for the donepezil group.
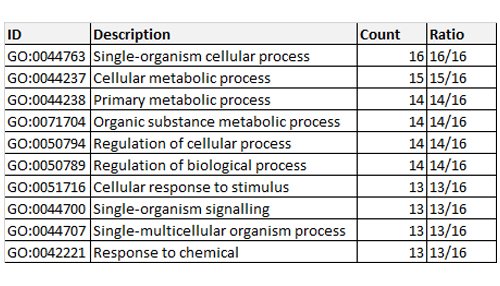

Table 1: Gene ontology enrichment for biological process associated with donepezil and its interacting proteins.
We conducted our programming and database integration with the R language, using the RStudio programming environment on an Intel Xenon CPU, 64-bit with dual processors (3.2GHz) and 128 GB of RAM. The R code was not compiled or optimised. R can be considered as the new de facto standard tool used in statistical research. R is highly versatile and highly expandable; over 5,000 packages have been developed by the highly active R community of researchers and developers.
The search for common side effects
Searching the SIDER2 database for drugs with common side effects that had a similarity score of 10% or above revealed 42 drugs were likely candidates. However, we only examined the top few in detail: aripiprazole, olanzapine, citalopram, fluoxetine, paroxetine, pregabalin, ropinirole and naltrexone. These drugs are all currently used to treat cognitive-related disorders. This could signify the network-based approach, using the side effects, could be a potential measure for establishing the common target sites between drugs. Some of these candidate drugs are currently prescribed to treat behaviour-related symptoms of Alzheimer’s, thus confirming our side effect hypothesis.
Here, we will only describe three drugs in detail. Aripiprazole has been tested as a drug to treat psychosis caused by Alzheimer’s. The research was evaluated in several clinical studies, which found that both fixed and flexible doses of the drug led to significant improvement in psychotic behaviour. Thus the evidence so far suggests aripiprazole could be a potential drug to treat this indication. However, some experts argue that the drug only resolves a few symptoms such as physiological symptoms, agitation and anxiety. The effective dose recommended in studies was 10mg/day, which has shown to improve physiological and behavioural symptoms. A common side effect experienced by the patients was found to be somnolence, although the extent of this was mild and moderate.
Olanzapine is an antipsychotic and also used to treat bipolar disorder. Low doses of olanzapine have shown to improve the behavioural symptoms caused by Alzheimer’s-related psychosis. As the dose of the drug is increased, the prevalence of somnolence in patients increased. In small doses it is well tolerated, however, olanzapine has high affinity binding for muscarinic receptor, which could result in cognitive impairing if it is administered over a longer time period or at a higher dose.
Research has found that selective serotonin re-uptake inhibitor (SSRI) drugs, which boost the level of serotonin signalling in the brain in order to treat depression, can reduce cognitive decline caused by Alzheimer’s. And so, the SSRI fluoxetine is now prescribed to counteract depression triggered by Alzheimer’s. The increased levels of serotonin correlate with a reduction in the production of beta-amyloid. This improves the cognitive functions of Alzheimer’s sufferers, and reduces any further decline in their condition. The increased level of serotonin also increases the alpha secretase enzyme, which is able to cleave the amyloid precursor at a different site than the one that results in the production of beta-amyloid.
Paroxetine, another SSRI, was previously prescribed to treat Alzheimer’s-related depression. It was stopped for this use when it was found to have anti-cholinergic character, thereby blocking the neurotransmitter acetylcholine (acetylcholine production increases blood flow in the brain; reduction of blood flow to the brain is associated with Alzheimer’s).
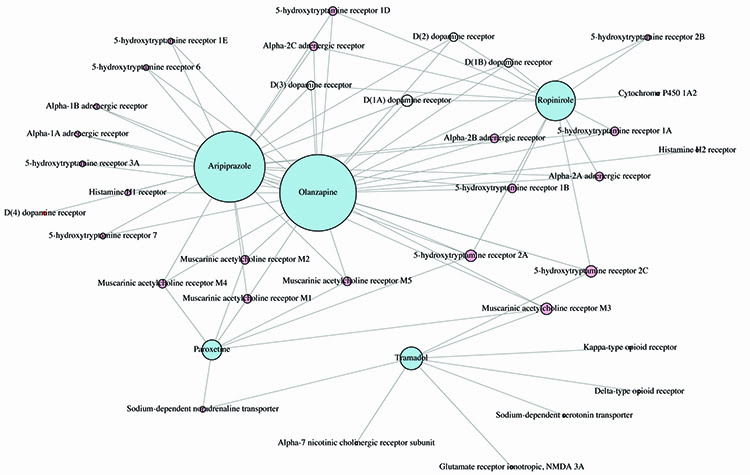

Figure 3: Complex graph network of the inter-connectivity patterns of protein activity for five candidate drugs; the node size is related to the number of shared targets.
Having identified our candidate drugs we constructed protein-to-protein interaction networks to seek a better understanding of the side effects and their intended targets. Figure 3 shows the network diagram for five of the candidate drugs. As can be seen they share a number of receptors and targets but enough are unique to each drug. This illustrates that not all SSRIs have the same mechanism of action in the body. This also implies that interaction with different targets can result in the same response. Paroxetine and fluoxetine are both SSRIs, but have a different mechanism of action.
Conclusions
We have demonstrated a very simple approach to drug repositioning by identifying common side effects to be used as a first pass of generating candidate drugs for further evaluation. We have demonstrated the usefulness of the side effect approach to screen candidate drugs for repositioning and the use of ontologies such as DO and GO provide a level of biological explanation. The ultimate test of course is have these drugs tested, at least possibly via in vitro means. Our future work will further investigate the chemical properties of the candidate drugs, integrating this information into a graphical network.
Biographies






References
- M Campillos, M Khun, A Gavin, L Jensen, P Bork. Drug target identification using side-effect similarity. Science. 2008; 321:263–266
- A Barabasi and Z Oltvai. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004; 5:101–113, 2004
- J Overington, B AlLazikani, A Hopkins. How many drug targets are there? Nature Reviews Drug Discovery. 2006; 5:993–996
- Lynn Schriml, Cesare Arze, Suvarna Nadendla, Yu-Wei Chang, Mark Mazaitis, Victor Felix, Gang Feng, and Warren Kibbe. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Research. 2012; 40:D940 – D946
- Jiang Li, Binsheng Gong, Xi Chen, Tao Liu, Chao Wu, Fan Zhang, Chunquan Li, Xiang Li, Shaoqi Rao, and Xia Li. Dosim: An Rpackage for similarity between diseases based on disease ontology. BMC Bioinformatics. 2011; 12(1):266
- K Shameer, B Readhead, J Dudley. Computational and experimental advances in drug repositioning for accelerated therapeutic stratification. Current Topics in Medicinal Chemistry. 2015; 15(1):5–20, 2015
- Jihong Yang, Zheng Li, Xiaohui Fan, Yiyu Cheng. Drug disease association and drug-repositioning predictions in complex diseases using causal inference probabilistic matrix factorization. Journal of Chemical Information and Modelling. 2014; 54(9):2562–2569, 2014
- P Imming, C Sinning, A Meyer. Drugs, their targets and the nature and number of drug targets. Nature Reviews Drug Discovery. 2006; 5:821–834, 2006
- M Kuhn, M Al Banchaabouchi, M Campillos, L Jensen, C Gross, A Gavin, P Bork. Systematic identification of proteins that elicit drug side effects. Molecular Systems Biology. 2013; 9(1)
- M Re and G. Valentini. Network-based drug ranking and repositioning with respect to drugbank therapeutic categories. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2013; 10(6):1359–1371
Related topics
Bioinformatics, Drug Development, Protein, Therapeutics, toxicology


