The frightening emergence of antibiotic-resistant bacteria
Posted: 17 February 2016 | Jean-Marie Frere (CIP Universite de Liege), Sebastien Rigali (CIP Universite de Liege) | 1 comment
Bacterial resistance to antibiotics is on the rise and a return to the ‘pre-antibiotic’ era has become a frightening possibility. Various factors are responsible for this situation: the overuse of antibiotics in human medicine and animal husbandry, insufficient public funding for research into fundamental bacteriology including resistance mechanisms, and a decreasing interest by the pharmaceutical industry in a field that is not considered to be sufficiently lucrative…

In this review, it is shown that the enzymes involved in peptidoglycan biosynthesis remain very attractive targets for new potential antibacterial compounds. The beta-lactam family of antibiotics and the glycopeptides (vancomycin) have been chosen as examples to illustrate resistance mechanisms. Meanwhile, it is a priority to better explore the Actinomycetes world, the source of currently-used biologically active compounds, for new targets. We know that there is a reservoir of such molecules, the scope of which has been underestimated, and many cryptic secondary metabolites might present antibacterial properties or inhibit antibiotic-degrading enzymes.
The emergence of antibiotic-resistant bacterial strains is not a new phenomenon. However, at the present time, large numbers of bacteria have become resistant to last-resort compounds such as carbapenems and colistin, while very few new compounds appear to be in development. As already mentioned by Caroline Richards in her editorial in the 2015 winter issue of Drug Target Review, the UK Prime Minister has appointed a commission, presided over by the economist Jim O’Neill, to examine the dire consequences of the continuing appearance of antimicrobial resistant (AMR) strains if efforts to counter this phenomenon are not significantly increased.
The predicted consequences are really frightening. Thirty-five years from now (in 2050), infections by resistant bacteria could be responsible for an additional 8,500,000 deaths per year, more than those due to all combined cancer forms. In addition to this human burden, the economic costs would be enormous: a cumulated impact of 100 trillion US dollars on the global world GDP is forecasted, resulting from premature deaths, absenteeism and the additional burden on social security systems resulting from increased durations of hospitalisation. There is thus an urgent need to analyse the causes of AMR and to propose effective counter-measures1. Indeed, the spread of resistant strains constitutes a major public health problem, not only in the fight against bacterial infectious diseases in the community, but also for various advanced hospital practices such as invasive surgery, organ transplant and cancer chemotherapy.
Why resistant strains emerge
The causes of the continuous emergence of resistant bacterial strains are now well established. In human medicine, the misuse or overuse of antibiotics is a determining factor2. For instance, it is known that viruses are responsible for the majority (up to 70%) of upper respiratory tract infections and that as a consequence the utilisation of antibiotics is useless in these cases. In many countries, over-the-counter sale of antibiotics without any medical prescription is a current practice, again resulting in the misuse and overuse of active compounds. In addition, many falsified drugs containing insufficient amounts of active principle are often marketed. In the veterinary field, the situation is even worse. In addition to purely therapeutic practices, the utilisation of antibiotics is widespread in intensive animal husbandry both for prophylactic reasons (note that the distinction between therapeutic and prophylactic utilisations is not always clear) and as growth promoters. The latter practice is now forbidden in the EU but not in other countries such as the USA and many developing nations, in particular the BRICS countries.
Although the general utilisation of antibiotics in animal husbandry has recently declined in the EU, it is on the rise in the rest of the world and expected to double globally over the next 20 years3,4. For instance, the reader is referred to a recent paper on the correlation between poultry meat and human infections by Salmonella5. Moreover, since many antibiotics are not metabolised by the human (or animal) body, they are rapidly excreted intact and pollute the environment. In consequence, wastewater treatment plants and surface waters become enriched with resistant strains, as shown by the prevalence of strains producing the NDM-1 beta-lactamase in India6.
A very god analysis of the environmental spread of antibiotic molecules and antibiotic-resistant bacteria and genes is given in reference4. Surprisingly, the AMR problem has received little attention from funding institutions. The O’Neill commission reports that, in the 2006-2010 period, this subject has only received 1.6% of all the NIH grant money, 15 and eight times less than cancer and AIDS research, respectively. In the EU, cancer research also receives much more grant money than AMR. The UK is probably the country where, proportionally, the largest amount of money is devoted to AMR research, but this is only 1% of the total research funds distributed by the Four Research Councils7. As a result, the best research groups have little incentive to centre their activities on the field and the best graduate students and post-docs tend to choose more ‘fashionable’ and better financed orientations.
In a similar way, many pharmaceutical companies have abandoned or spun-off antibacterial research and development so that very few new druggable compounds have been produced over the past few years. The main reasons for this lack of interest appear to be purely financial since much better returns are expected from compounds designed to treat chronic diseases (diabetes, hypertension) or cancer. One can cynically state that antibiotics development constitute a huge financial drawback: they cure the patient after about 10 days (or the patient dies!) in contrast to chronic diseases where the treatment might last for months, years or the rest of the patient’s life. However, it is not all doom and gloom: avibactam (AstraZeneca) and BAL 29880 (Basilea) have recently been developed and exhibit very interesting properties as beta-lactamase inhibitors.
In the following paragraphs, we develop the story of beta-lactams that remain, by far, the most widely utilised antibacterial compounds and which represent a striking example of the continuous fight between the scientists and the pathogenic bacteria.
Beta-lactams
The first mention of “an enzyme from bacteria able to destroy penicillin” pre-dated the determination of the exact structure of the antibiotic8. The first major success of penicillin (benzylpenicillin, Figure 1A) was in the fight against staphylococcal infections, first on the battlefield at the end of World War II, and later in hospitals. Rapidly, resistant strains emerged that produced a penicillin-destroying enzyme called penicillinase. After a few years, these strains represented a majority of those isolated in the London hospitals. The problem was solved by modifying the C-6 penicillin side chain (methicillin, Figure 1B) and the discovery of cephalosporins (Figure 1D). Methicillin and cephalosporins escaped the activity of the staphylococcal beta-lactamase.
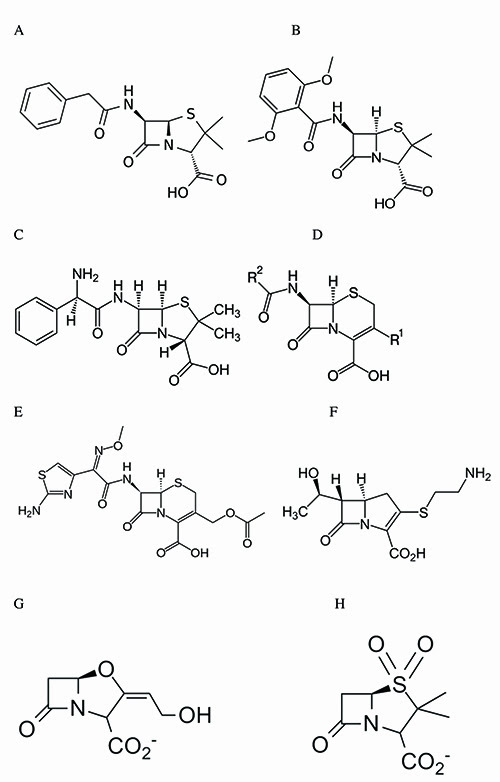

Figure 1: Structures of some beta-lactams. A: benzylpenicillin (penicillin G); B: methicillin; C: ampicillin; D: general structure of cephalosporins; E: cefotaxime; F: imipenen (a carbapenem); G and H: beta-lactamase inactivators (clavulanate, G and sulbactam, H).
Another modification of the penicillin side-chain yielded ampicillin (Figure 1C), which was much more effective than benzylpenicillin against Gram-negative strains. These successes resulted in some complacency, as shown by this statement of Gen. William H Stewart, US Surgeon General in 1969: “The time has come to close the book on bacterial diseases”. But meanwhile, a plasmid-borne enzyme was detected that could hydrolyse many penicillins and first generation cephalosporins: the TEM beta-lactamase. At about the same time, the appearance of methicillin-resistant staphylococcal strains was reported (the so-called methicillin-resistant Staphylococcus aureus, or MRSA) whose resistance was not due to the production of a beta-lactamase but to the acquisition of a resistant penicillin-binding protein (PBP).
Indeed, the antibacterial properties of beta-lactams rests on the fact that they acylate the hydroxyl group of the active-site serine residue9 of the D-Ala-D-Ala-transpeptidases that are responsible for the formation of the peptide cross-links between the glycan strands of the peptidoglycan and are thus essential to the survival of bacteria in a hypotonic environment. These enzymes are usually referred to as PBPs. All bacterial cells contain a set of different PBPs that display different functions in peptidoglycan metabolism, but a complete analysis of these functions is beyond the scope of the present short review (for more details on beta-lactamases and PBPs, see references 10 and 11).
The sensitivity of a PBP to a particular beta-lactam depends on the rate of the acylation reaction. Resistance at the PBP level is due to low-to-very-low acylation rates and is mainly encountered in Gram-positive bacteria. The origin of these ‘resistant’ PBPs varies according to the bacterial species (or genus). MRSA has acquired a new gene encoding PBP2a from another naturally resistant staphylococcal species, probably Staphylococcus sciuri. In Streptococcus pneumoniae, the resistant PBPs are encoded by ‘mosaic genes’ that contain portions of an intrinsic gene and of imported genes that code respectively for a sensitive and more resistant PBP. Finally, Enterococci produce an intrinsic resistant PBP but in amounts insufficient to ensure a sufficient level of cross-linking. Resistance arises from overproduction of the resistant PBP due to deregulation of the transcription of the corresponding gene and can be further increased by point mutations.
Most of the Gram-positive resistant strains remain sensitive to vancomycin, an antibiotic that binds to the D-Ala-D-Ala C-terminus of the stem peptide and, in consequence, also inhibits the transpeptidation reaction. Resistance to vancomycin has also been observed (resulting from the replacement of the D-Ala-D-Ala C‑terminus of the stem peptide by D-Ala-D-lactate) but this is another story.
It is interesting to note that PBP-mediated acquired resistance in pathogens is mainly restricted to Gram-positive bacteria that do not produce beta-lactamases. Staphylococcus aureus is clearly the exception but for some unknown reason, its ‘penicillinase’ never evolved into a broad spectrum beta-lactamase. In Gram-negative species, the production of beta-lactamases sometimes accompanied by a low or decreased outer membrane permeability and/or by the overexpression of efflux systems constitutes the major resistance factor. As mentioned above, the emergence of plasmid-borne beta-lactamases such as TEM and SHV was followed by their rapid spreading to several pathogenic species and resulted in a search for beta-lactamase-resistant cephalosporins and for beta-lactamase-inactivating compounds. Both approaches were successful, with the discovery of compounds such as cefotaxime (Figure 1E) and ceftazidime that were very poorly hydrolysed by the original TEM and SHV enzymes and of clavulanate (Figure 1G), sulbactam (Figure 1H) and, much later, avibactam that inactivated these enzymes effectively.
However, the sometimes improper utilisation of cefotaxime and ceftazidime resulted in the selection of extended-spectrum beta-lactamases (ESBLs). Some of these were derived from the original TEM and SHV enzymes thanks to a few mutations in the part of the active-site pocket where the R2 side-chain (Figure 1D) of the cephalosporins binds. Others were new enzymes, exhibiting only 30-40% sequence similarity to TEM or SHV. Moreover, in Enterobacteriaceae, the deregulated synthesis of chromosome-encoded class C beta-lactamases resulted in very high periplasmic enzyme concentrations so that the MIC values increased significantly, even for very poor substrates12. On the other hand, inhibitor-resistant TEM derivatives were also detected but the increase in resistance remained modest when compared with the increased activity of other variants against cefotaxime and ceftazidime.
In the early 1990s, carbapenems (Figure 1F) were introduced into the antibacterial arsenal. In the beginning, they appeared to be ideal compounds: they reacted rapidly with most PBPs and were quite resistant to nearly all known beta-lactamases; they rapidly became considered as last-resort antibiotics. But new beta-lactamases that could hydrolyse them were also selected. Some belonged to the ‘classical’ active-site serine groups (e.g., the KPC family) but others were members of a completely different group: the metallo-beta-lactamases containing one or two catalytic Zn++ ions (MBLs). Such enzymes had been known for a long time but they were produced by relatively innocuous Bacillus cereus strains and rather considered as biochemical curiosities. However, plasmid-borne MBLs were detected in the 1990s and later, the IMP and VIM and, more recently NDM families.
The latter is particularly worrying, not because NDM is an ‘exceptional’ enzyme as was claimed by some authors but because the corresponding gene is carried by a plasmid encoding factors responsible for resistance to other families of antibiotics. The MBL genes have spread rather rapidly worldwide. These enzymes are a major problem since they hydrolyse all classes of beta-lactams with the sole exception of monobactams (e.g., aztreonam) and are, not surprisingly, insensitive to the available beta-lactamase inactivators. To complicate the situation, some bacterial strains produce several beta-lactamases, for instance an MBL and an active-site serine enzyme, so that an aztreonam-avibactam combination is presently under clinical trial.
Colistin: an endangered last hope?
Colistin is a mixture of two polypeptides: polymyxins A and B. It acts by perturbing the structure of the outer membrane and has a detergent-like effect on the cytoplasmic membrane. It has been known since 1949, but was not widely used in human medicine due to unfavourable side effects (nephro- and neuro-toxicity). Recently, it was revived as a last-resort compound in the fight against multi-drug resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumanii and NDM-producing Enterobacteriaceae, however, several other species are not sensitive.

In the last few years, resistance due to the presence of the transmissible mcr-1 gene has started to appear13. It seems that this emergence is due, at least in part, to the widespread utilisation of colistin in agriculture and many authors have suggested that the agricultural use of this antibiotic should be strongly, if not completely curtailed.
Peptidoglycan biosynthesis as an antibiotic target
Peptidoglycan is a specific and essential bacterial polymer. It contains D-amino acids that are rare in mammals. This is the main reason why it remains a particularly interesting target for the development of new antibiotics. The last two steps in its biosynthesis (the glycosyltransferase and transpeptidase reactions) present the additional advantage of taking place at the outer surface of the cytoplasmic membrane. This means that an antibiotic molecule does not encounter any permeability barrier in Gram-positive bacteria (with the exception of Corynebacterineae that include mycobacteria) and only have to diffuse through the outer membrane in the case of Gram-negatives and mycobacteria. Consequently, the problem of crossing the cytoplasmic membrane is avoided. These two factors, specificity and easy access to the target enzymes, explain the success of beta-lactam antibiotics which results in great part from the absence of secondary effects, if one excepts a few cases of allergy that will not be discussed here.
Hopes versus future concerns
The alarming assessment that human pathogens are currently winning the battle against our old antibiotics is making the return to the pre-antibiotic era a realistic scenario. Decades of misuse of these medicines have rendered our natural enemies (i.e., bacteria) less vulnerable. However, hope is again arising from…bacteria. More specifically, this wave of optimism relies in part on studies of Streptomyces and other actinobacteria called by David Hopwood the ‘antibiotic makers’ since they have offered us most of the natural compounds currently used in human therapy14.
In 2002, publication of the genome of the model species Streptomyces coelicolor15 revealed the presence of the so-called ‘cryptic’ (hidden, orphan) antibiotics. These compounds have not been identified yet mainly because expression of their biosynthetic genes is silent under laboratory growth/culture conditions. Importantly, S. coelicolor is not an isolated case and all the genomes of filamentous actinobacteria reveal high potential to produce novel compounds suggesting that we have so far just picked the low-hanging fruits. Discovering what controls cryptic secondary metabolite production is guaranteed to result in novel antibiotics being found. In addition, research programs aimed at isolating novel actinobacteria residing in unexplored habitats are no longer considered ‘unfashionable pursuits’ and many novel species are being discovered every year.
In addition, sequencing the genomes of these novel species identifies gene clusters potentially involved in the biosynthesis of new compounds that might exhibit antimicrobial activity16. The genetic information for the production of future antibiotics is probably already known and the genes of resistance to these unknown antibiotics also already exist. Indeed, being able to resist an antibacterial agent is a prerequisite for the producing microorganism in order to avoid suicide. If actinobacteria have, on the one hand, provided antibiotics, then on the other hand, they have also contributed to the spread of resistance genes and acquisition of these genes by pathogens17,18. The forthcoming generation of natural antibiotics can give us a ‘second chance’ to win the fight against our natural enemies. But the battle is already lost if we ignore the causes of the current dramatic situation.
Hopefully we now have a unique opportunity to show that we can learn from our past mistakes. In this respect, it is evident that the funding of fundamental bacteriology should be very strongly increased, both to support the discovery of new compounds and to improve our knowledge of the genetics, biochemistry and physiology of the pathogens. In addition, it is necessary to promote more responsible and specific use of the present and future antimicrobial arsenal. Indeed, antibiotics are known to perturb the intestinal flora, which can result in many
unexpected pathologies19. Rapid identification of the infectious agent and of its characteristics, followed by administration of a very specific antibiotic should minimise these perturbations and, in addition, limit the selection of resistant strains of innocuous (or even beneficial) bacteria. In a timely manner, Philippe Villain-Guillot mentioned recently in this review that it was “time to renew the antibiotic arsenal”20. As suggested by Woolhouse and Farrar21, an international panel similar to that involved in the analysis of climate change (IPCC) should be appointed to, for instance, limit or eliminate the utilisation of antibiotics in agriculture where this practice is justified only on economic grounds. Are immediate economic gains more important than the health of future generations?
Acknowledgements: the authors wish to thank Professor Roger Pain for his excellent suggestions.
Biographies




References
- To access the full reports of the O’Neill Commission, just type “Review on Antimicrobial Resistance” in your search engine
- Reardon S. http://nature.com/new/dramatic-rise-seen-in-antibiotic-use-1.18383
- Van Boeckel TP et al. Global trends in antimicrobial use in food animals. PNAS (2015) Vol 112, pp5649-5654
- Silvera MA, Freitas AR, Peixe L. and Novais C. Environmental spread of antibiotic molecules, antibiotic resistant bacteria and genes: jigsaw pieces of a public health problem. Revista da Faculdade de Ciências da Saude, Porto. (2009) Vol 6, pp 244-253
- Antunes P, Mourao J, Campos J and Peixe L. Salmonellosis : the role of poultry meat. Clinical Microbiology and Infection. (2016) doi: 10.1016/j.cmi.2015.12.004
- Kumarasamy KK et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. (2010) Vol 10 pp 597-602.
- Kelly R, Zoubiane G, Walsh D, Ward R and Goossens H. Public funding for research on antibacterial resistance in the JPIAMR countries, the European Commission and related European Union agencies: a systematic observational analysis. The Lancet Infectious diseases. doi 10.1016/S1473-3099(15)00350-3 (published online Dec 18, 2015)
- Abraham EP and Chain E. An enzyme from bacteria able to destroy penicillin. Nature (1940) Vol 146, p 837
- Frère JM, Duez C, Ghuysen and J Vandekerckhove. Occurrence of a serine residue in the penicillin-binding site of the exocellular DD-carboxypeptidase-transpeptidase from Streptomyces FEBS Lett. (1976) Vol 70 pp 257-260
- Frère JM, Sauvage E and Kerff F. From “an enzyme able to destroy penicillin” to carbapenemases: 70 years of beta-lactamase misbehavior. Current Drug Targets (2016) Vol 17, in press
- Sauvage E, Kerff F and Terrak M. The penicillin binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev (2008) Vol 32 pp 234-258
- Lakaye B, Dubus A, Lepage S, Groslambert S and Frère JM. When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Moleculat Microbiology (1999) Vol 31 pp89-101
- Liu YY et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious diseases. doi 10.1016/S1473-3099(15)00424-7 (published online Nov 18, 2015). See also the numerous comments in later online editions of the same journal
- Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford University Press (2007). https://books.google.be/books?hl=fr&lr=&id=IO9cCAAAQBAJ&oi=fnd&pg=PT10&dq=david+hopwood+nature+medicine&ots=UMmG1FpHsW&sig=U6T-m8jd4KY-k5sgTxleZK_6_gs
- Bentley SD et al. Complete Genome Sequence of the Model Actinomycete Streptomyces Coelicolor A3(2). Nature (2002) Vol 417, pp 141–47
- Kolter R and van Wezel GP. Goodbye to brute force in antibiotic discovery? (2016). Nature Reviews Microbiology, February 2016 Doi:10.1038/nmicrobiol.2015.20
- D’Costa VM, McGrann KM, Hughes DW and Wright GD. Sampling the Antibiotic Resistome’. Science (2006) Vol 311, pp 374–77
- Davies J and Davies D. Origins and Evolution of Antibiotic Resistance’. Microbiology and Molecular Biology Reviews (2010) Vol 74 pp 417–33
- Schulfer A and Blaser MJ. Risks of antibiotic exposures early in life on the developing microbiome. PloS Pathog (2015) Vol 11: e1004903. doi: 10.1371/journal.ppat.1004903. eCollection 2015 Jul. Prof Blaser has published many other papers on the correlations between various diseases and antibiotic overuse
- Villain-Guillot P. Time to renew our antibiotic arsenal. Drug Target review (2016) January 19
- Woolhouse M and Farrar J. An intergovernmental panel on antimicrobial resistance. Nature (2014) Vol 509, pp555-557
Related topics
Antibiotics, Pathology & Molecular Medicine, Therapeutics



I don’t even know the way I finished up right here,
however I thought this put up was good. I don’t realize who you might be however definitely you’re going to a famous blogger should you aren’t already.
Cheers!