NGS: empowering infectious disease research beyond reality
Posted: 13 September 2017 | Pooja Sharma, Pushpanathan Muthuirulan | No comments yet
Pushpanathan Muthuirulan & Pooja Sharma explain how high-throughput next-generation sequencing (NGS) technologies are revolutionising our ability to characterise infectious diseases at genomic, transcriptomic and epigenetic levels…


NGS technology can be used to describe the microbiome in health and diseases states, and it has helped to identify virulence genes for pathogenicity, driven insights into genetic difference among related pathogens, enabled development of diagnostic tools for discrimination among specific strains, revealed mechanisms of host resistance, and also provided comprehensive understanding of host-microbe interactions in infectious disease progression. This article discusses the benefits and challenges of using NGS in infectious disease diagnosis.
Next-generation sequencing (NGS) technology has revolutionised the diagnosis and treatment of infectious diseases. The standard long-time approach of culture-based testing for the diagnosis of infectious diseases is still in use. However, this approach has several limitations including the requirement for dedicated trained staff and intrinsic inefficiency in the propagation of certain microbial species (eg, fastidious bacteria and viruses). Culture-based assays are often cost prohibitive, time consuming and inefficient.1 They have been progressively replaced by nucleic acid-based detection methods like PCR, which allows greater speed, low cost, sensitivity and specificity. The main drawback with a PCR-based method is its ability to identify only predefined or conserved targets of the pathogens, and it does not discriminate between genotypes which further limits detection of emerging pathogens.2 Several other methods such, as DNA hybridisation, multiplex PCR assays, enzyme immunoassays, and pulsed-field gel electrophoresis, have been developed to broaden the specificity and sensitivity of pathogen detection. However, these methods have not proven satisfactory due to lack of sensitivity for clinical diagnosis.3
NGS has recently revolutionised the field of medical microbiology and it serves as an excellent diagnostic tool for infectious diseases. Technological advancement has overcome limitations in the timeliness as well as well as accuracy of pathogen identification, and it offers incredible opportunities for cost-effective and efficient ways for addressing medically relevant issues.4 The NGS technological platform can also capture target genomic regions of interest and allows massive parallel sequencing of those regions along with high-performance computing to identify genomic variants or signatures that may be biologically relevant. This remarkable technological advancement has generated huge amounts of data addressing medically significant issues, including clinical assessment of individuals with genetic disorders.5 Thus, NGS overcomes most hurdles of conventional single-gene testing that often end the ‘diagnostic odyssey’ of rare diseases.
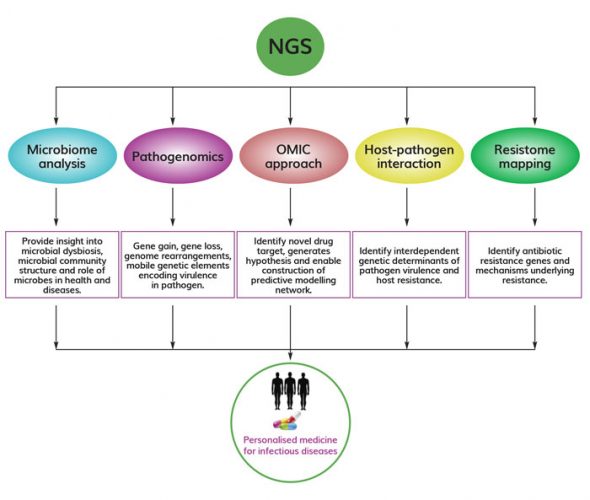

Figure 1. Versatile application of NGS in infectious disease research NGS has widespread application in infectious disease research, including microbiome analysis, pathogenomics, integration of omics approaches, host-pathogen interaction studies, and resistome mapping. All these approaches could aid in development and implementation of personalised medicine to treat infectious diseases.
Recent advances in NGS also allow researchers to identify uncultivable pathogens that might be missed in routine cultures and diagnostic sensitivity assays. It offers attractive ways to distinguish between different pathogen strains, detect co-infections and uncover emerging pathogens. Whole genome sequencing (WGS) and RNA sequencing using NGS have uncovered new genomic sequences and functional information that has been used successfully to uncover mutations associated with antibiotic resistance and the spread of drug-resistant pathogens.6 Thus it has become the gold-standard method on time and is now being considered as a routine testing option by many clinical researchers to solve the mystery behind the cause of infectious diseases. This article mainly covers the potential application of NGS in infectious disease research, including microbiome disease association, pathogenomics, integrative omics approaches for predictive modelling of infectious disease, host-pathogen interaction and resistome mapping that would allow infectious disease professionals or clinicians to identify, track and respond to disease outbreak by developing novel therapeutics (Figure 1).
Microbiome analysis
The advent of NGS has enabled high-throughput profiling of microbiomes that has allowed researchers to better understand the role of microbes in health and diseases.7 It has also often provided deeper insight into acute or chronic infectious diseases involving microbial dysbiosis.8 Traditional methods of microbial characterisation in clinical or environmental samples relied on labour-intensive microbiology techniques such as microbial enrichment and isolation of individual organisms followed by phenotypic and genotypic analysis.9 Microbial community profiling within a single sample was not easily achievable using traditional techniques, but NGS has enabled high-throughput microbial community profiling of multiple samples simultaneously with less cost and much faster speed.10 The cost-effective and high-throughput sequencing capacity of NGS has fueled metagenomics studies that have surveyed targeted genomic regions of entire microbial communities, including those of uncultivable organisms. The two main approaches of microbiome analysis, such as 16S/23S ribosomal RNA (rRNA) amplicons and shotgun metagenomics, enabled investigation of the microbiome and inference of the microbial community’s metabolic abilities with high throughput and unprecedented resolution.11 NGS offers sophisticated technological platforms that can characterise the structure of microbial communities and their metabolic capabilities around infectious diseases. This can help researchers to identify new therapeutic targets and develop novel approaches to treating infectious diseases.
Pathogenomics
NGS has been widely used in clinical laboratories to discover novel pathogens. Unlike other diagnostic assays, NGS requires little or no prior knowledge of pathogen, giving it an impressive advantage over other diagnostic assays.12 The field of ‘pathogenomics’ has uncovered the emerging and re-emerging microbial pathogens that translated genomic technologies into a surveillance and diagnostics method. Pathogenomics has enabled high-resolution mapping of the genetic determinants in microbes that underpin pathogenicity.13 Whole genome sequencing (WGS) has emerged as the preferred technology for analysing the dynamic genomes with a high plasticity that are needed for pathogens to survive in challenging environments.14 The influx of large data sets generated by WGS have been used to detect, compare and catalogue dynamic regions in the genome that lead to pathogen development. It has also provided insight into microbial evolutionary genomic events that shape pathogen genomes, such as gene gain, gene loss and genome rearrangement.15
Comparative approaches have uncovered the pan genome (entire gene pool of pathogen species that includes genes not shared by all strains) and core genome (set of genes found across all strains of a pathogen species) of different pathogen species and enabled researchers to determine the variability of different strains as a mechanism of survival and virulence.16 Bioinformatics efforts in pathogenomics have also helped to identify mobile genetic elements that encode virulence factors, such as plasmids, pathogenicity islands, and prophages.17 Thus, NGS technology sheds light on unique dynamic regions of pathogens’ genome that opened up new possibilities to explore those regions as novel therapeutic targets for infectious diseases.
Omics approaches
Systems biology approaches have been implemented to overcome the obstacles in infectious disease research. Mathematical models of complex biological processes have provided a basic framework for the understanding of dynamic interaction and cross talks between host and pathogen.18 NGS technology has opened the door to sequencing total transcriptomes that dictate host responses to microbial infections. Proteomic and metabolomic technologies are evolving rapidly with greater throughput and sensitivity. Integration of high-throughput multi-omics data (transcriptomics, proteomics and metabolomics) offers the most effective way to address the challenges of infectious disease research.19
Omic approaches have helped to construct a predictive model of the network and infer complex dynamic interactions between host and pathogen. Omic strategies involve iterative rounds of model development, testing, and filling in knowledge gaps with experimental data. The comprehensive predictive models generated based on experimental data can be used to identify potential therapeutic targets to combat infectious diseases. Omic investigations are increasingly being used to understand the aetiology of infectious diseases, biomarker discovery, drug discovery and toxicity assessment.20 Thus, these approaches have proved helpful in predictive modelling of infectious disease networks, identifying novel drug targets, generating hypotheses, and predicting the cause and outcome of infectious diseases.
Host-pathogen interaction
The development of NGS technologies has opened up new molecular perspectives that allow a deeper understanding of host-pathogen interaction at a precise molecular level. The influx of large data sets being generated from genomic and transcriptomic approaches has enabled researchers to characterise the molecular forces driving host-pathogen interactions. During recent years, NGS technologies have unravelled the genomic basis of adaptation that allows interdependent genetic determinants of pathogen virulence and host resistance to be identified.21 It is increasingly being used to identify virulence factors, detect sequence variations (single nucleotide polymorphism, insertion and deletions) and transfer of genetic materials between pathogen and host.22
Further, NGS technologies have been used to decipher the complex temporal and spatial dynamics of host-pathogen interaction toward understanding the host-response pathways during the disease process. Transcriptomic approaches have characterised the expressed portion of the genome that effectively opened up possibilities to measure the transcriptomic response of host and pathogens during infection.23 Thus, genomic and transcriptomic approaches have provided insight into genome-wide interaction between host and pathogen, enabling researchers to understand the precise molecular mechanisms involved.
Resistome fingerprinting
Antibiotic resistance has become a major threat to global health. Understanding the molecular diversity or resistome composition underpinning resistance would guide efforts to develop new antibiotics for individualised treatment. Rapid resistome profiling using an NGS-based functional-metagenomics approach would be an effective approach towards personalised antibiotic treatment. During recent years, NGS has been successfully used to characterise the gut resistome of intensive care unit (ICU) patients using metagenomic functional selection.24
NGS-based metagenomics studies have mapped the human gut resistome with unprecedented resolution and shown that anaerobic gut commensal organisms are the main reservoir of antibiotic resistant genes.25 However, many questions relating to the transfer of antibiotic resistance genes from harmless gut commensals to opportunistic pathogens remain unanswered. To fully assess the risk of antibiotic-resistance genes, further studies are highly warranted to completely profile antibiotic-resistance genes with respect to their potential for horizontal gene transfer. Thus, NGS-based mapping of the resistome could aid personalised antibiotic therapy in high-risk patients.
To conclude, NGS is the most powerful method that allows molecular detection and genotyping of infectious disease pathogens. NGS can detect uncultivable pathogens, distinguish different strains of pathogens, and even uncover emerging pathogens. NGS offers an attractive high-throughput technological platform for microbial identification, mapping genetic determinants underpinning pathogenicity, construction of predictive models for infectious diseases, and understanding cross-talks between host-pathogen interactions and resistome mapping. Thus, NGS has emerged as a gold standard method for infectious diseases research, enabling clinicians and research professionals to develop efficient diagnostic tools and therapeutic approaches to treating infectious diseases.
Author Biographies


Pushpanathan Muthuirulan is a postdoctoral researcher at the National Institutes of Health, working in the field of neuroscience, where his research focuses on developing tools to map complex neural circuits. He is a biologist with eight years of research experience in the discovery and development of drug candidates. Dr Muthuirulan has a BSc and MSc in Zoology from the American College and a PhD in Microbiology from Madurai Kamaraj University
Address for correspondence
Laboratory of Gene Regulation and Development,
National Institute of Child Health and Human Development,
National Institutes of Health, Bethesda, MD 20892, USA
Phone: +1301-674-3108,
Email: pushpanathan31@gmail.com; pushpanathan.muthuirulan@nih.gov 

Pooja Sharma is a Faculty Assistant at the University of Maryland and US Department of Agriculture, where her research has focused on elucidating the molecular biology and immunology of host-parasite interactions in model animals and domestic livestock.
References
- Lindsay B, Pop M, Antonio M, Walker AW, Mai V, Ahmed D, et al. Alternative methods of bacterial pathogen detection: culture, GoldenGate®, Universal Biosensor®, 16S rRNA-Gene Survey. Clin. Microbiol. 2013;51:3263–9.
- Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Cell. Infect. Microbiol. 2014;4:25.
- Dabisch-Ruthe M, Vollmer T, Adams O, Knabbe C, Dreier J. Comparison of three multiplex PCR assays for the detection of respiratory viral infections: evaluation of xTAG respiratory virus panel fast assay, RespiFinder 19 assay and RespiFinder SMART 22 assay.BMC Infect. dis. 2012;12:163.
- Buermans HPJ, Den Dunnen JT. Next generation sequencing technology: advances and applications. BBA-Mol. Basis Dis. 2014;1842:1932-
- Schmidt B, Hildebrandt A. Next-generation sequencing: big data meets high performance computing. Drug Discov. Today. 2017;22:712-717.
- Burkholder WF, Newell EW, Poidinger M, Chen S, Fink K. Deep sequencing in infectious diseases: immune and pathogen repertoires for the improvement of patient outcomes. Immunol. 2017;8:593.
- Autenrieth IB. The microbiome in health and disease: a new role of microbes in molecular medicine. J. Mol. Med. 2017;95:1-3.
- Clooney AG, Fouhy F, Sleator RD, O’Driscoll A, Stanton C, Cotter PD, et al. Comparing apples and oranges?: next generation sequencing and its impact on microbiome analysis. PLoS One. 2016;11:e0148028.
- Ellis RJ, Morgan P, Weightman AJ, Fry JC. Cultivation-dependent and-independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Environ. Microbiol. 2003;69:3223-3230.
- Smith AM, Heisler LE, St Onge RP, Farias-Hesson E, Wallace IM, Bodeau J, et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38:e142-e142.
- Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Microbiol. 2016;7:459.
- Frey KG, Bishop-Lilly KA. Next-Generation Sequencing for Pathogen Detection and Identification. Methods Microbiol. 2015;42:525-
- Demuth A, Aharonowitz Y, Bachmann TT, Blum-OehlernG, Buchrieser C, Covacci A, et al. Pathogenomics: an updated European Research Agenda. Genet. Evol. 2008;8:386-393.
- Edwards RA, Rohwer F. Opinion: viral metagenomics. Rev. Microbiol. 2005;3:504.
- Fraser-Liggett CM. Insights on biology and evolution from microbial genome sequencing. Genome Res. 2005;15:1603-
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Opin. Microbiol. 2008;11:472-477.
- Pallen MJ, Wren BW. Bacterial pathogenomics. Nat. 2007;449:835.
- Baloni P, Ghosh S, Chandra N. Systems approaches to study infectious diseases. In Synth. Biol. 2015:(151–172) (Springer Netherlands).
- Aderem A, Adkins JN, Ansong C, Galagan J, Kaiser S, Korth MJ, et al. A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. MBio. 2011;2:e00325-10.
- Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. The Obstetrician & Gynaecologist. 2011;13:189-
- Greenwood JM, Ezquerra AL, Behrens S, Branca A, Mallet L. Current analysis of host–parasite interactions with a focus on next generation sequencing data. Zoology. 2016;119: 298-
- Schrider DR, Begun DJ, Hahn MW. Detecting highly differentiated copy-number variants from pooled population sequencing. In Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2013;344.
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. methods. 2008;5:621-628.
- van der Helm E, Imamovic L, Hashim Ellabaan MM, van Schaik W, Koza A, Sommer, MO. Rapid resistome mapping using nanopore sequencing. Acids Res. 2017;45:e61-e61.
- van Schaik W. The human gut resistome. Trans. R. Soc. B. 2015. 370:20140087.
The rest of this article is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- quarterly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Related topics
Next Generation Sequencing (NGS), Technology
Related organisations
Catholic University of America, National Institutes of Health (NIH)
Related people
Pooja Sharma, Pushpanathan Muthuirulan


