Researchers create new Cas9 variant to reduce off-target CRISPR edits
Posted: 4 March 2020 | Victoria Rees (Drug Target Review) | No comments yet
A study has shown that altering amino acid residues in Cas9 to produce new variants can produce a vector with increased gene editing specificity.

Researchers have developed a Cas9 variant that they report increases the specificity of CRISPR genome editing. The study was conducted at the Max Planck Unit for the Science of Pathogens and the Faculty of Medicine at the Martin Luther University Halle-Wittenberg, both in Germany.
Cas9 is able to target certain areas of DNA through guidance from RNA, which contains the complementary sequence to the DNA target site. “Sometimes, however, Cas9 can also cut DNA sequences that are very similar to the actual target, known as off-targets,” explained Emmanuelle Charpentier, director of the Max Planck Unit for the Science of Pathogens.
This undesired activity of CRISPR-Cas9 can lead to inaccuracies in genome editing: “An unintended cut at the wrong place in the human genome can have profound consequences. That is why we need a more specific system,” says Michael Böttcher, Assistant Professor at the Medical Faculty of the Martin Luther University.
In the current study, the team of researchers focused on an evolutionarily conserved domain of Cas9, known as bridge helix. They found that the bridge helix plays a critical role in the mechanism by which Cas9 interacts with its guide RNA and DNA target site. They identified a group of amino acid residues that make contact with the phosphate backbone of the guide RNA, thereby facilitating the formation of a stable loop, which is essential for the activity of Cas9. In such a loop, the Cas9-bound guide RNA pairs with the complementary strand of the DNA target sequence while displacing the second DNA strand, thereby both DNA strands are cut.
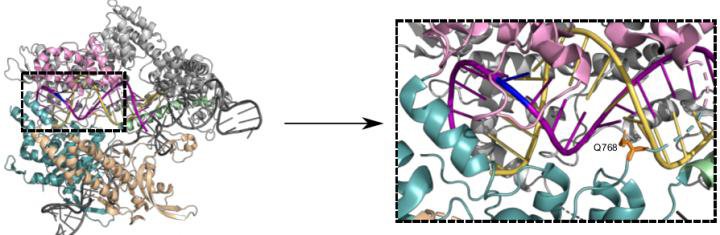

Structure of Cas9 bound to single guide RNA (scaffold in dark grey, spacer in yellow) and DNA target strand (purple). Changing the amino acid residue Q768 (orange, dashed box) yields a more specific Cas9 variant (credit: Bratovic et al., 2020).
The researchers generated new variants by changing these amino acid residues and found that several variants cut much less frequently at off-target sites than the original Cas9 enzyme. They further showed that one of the identified variants, called R63A/Q768A, increased the gene editing specificity of Cas9 in human cells too.
“Our results provide a new basis for further optimisation of CRISPR-Cas9. They demonstrate the need to gain more knowledge about the biochemistry of CRISPR-Cas systems to further improve them,” concluded Charpentier.
The study was published in Nature Chemical Biology.
Related topics
CRISPR, DNA, Drug Targets, Gene Therapy, Genome editing, Genomics, RNAs
Related organisations
Martin Luther University Halle-Wittenberg, Max Planck Unit for the Science of Pathogens
Related people
Emmanuelle Charpentier, Michael Böttcher


