The therapeutic potential of RNA directed epigenetic regulation of transcription
Posted: 10 May 2014 | Kevin V. Morris (University of New South Wales / The Scripps Research Institute) | No comments yet
Developments over the past decade have demonstrated that non-coding RNAs can exert control of transcriptional gene expression. Recent advances in genome-wide studies have revealed an abundance of long non-coding RNAs (lncRNAs) in human cells, which are as numerous as protein-coding genes…


Notably, some of these lncRNAs actively modulate gene transcription and epigenetic states. Mechanistically, some of these lncRNAs appear to recruit epigenetic silencing complexes to their homology containing loci. I present here details of this mechanism as well as insights that now allow us to mimic lncRNA function to regulate a genes expression, either epigenetically silencing or de-repressing and activating gene expression. These insights suggest that long-term heritable control of gene transcription can be instilled on target genes in a directed manner. Such insights portend an entirely new therapeutic platform for targeted gene regulation.
Long non-coding RNAs and gene expression
A profound realisation and subsequent paradigm shift is occurring in molecular and cellular biology. This shift in our collective thinking is the result of exciting findings from the ENCODE consortium. ENCODE, a consortium of over 280 scientists, determined that ~80 per cent or greater of the human genome is transcribed and possibly functionally active, despite lacking protein-coding capabilities1,2. These genome-wide observations resonate with what others and we have observed during the last decade, while studying non-coding RNA directed transcriptional gene regulation, and drastically expand on the inherent complexity of the cell (reviewed in3,4). Most notably are the recent observations that antisense long lncRNAs5 appear to be active transcriptional regulators of particular protein-coding genes6-9. Many of these regulatory antisense lncRNAs appear to emanate from pseudogenes, which were thought to be evolutionarily remnants in the human genome8,9.
Non-coding RNA directed discordant regulation
The first observations suggesting lncRNAs might be functional gene regulators were the result of the FANTOM consortium, where deep sequencing of around one per cent of the mouse transcriptome was carried out. In this body of work, it was observed that several genes exhibited transcription in both directionalities and that when some of these transcripts were perturbed, there was an effect on their respective sense or antisense counterparts5. Not much later observations in human cells, specifically carried out on the p21 and p15 tumour suppressor genes6,7, indicated that these genes were also under similar forms of RNA directed regulation as observed in the mouse10. In these studies it became apparent that transcription for these tumour suppressor genes was occurring in both directions. These bidirectional long antisense non-coding RNA (lncRNAs) were also found to be functional in epigenetically regulating the expression of their sense protein-coding gene counterparts (reviewed in11). Interestingly, a large percentage of those genes which have been found to exhibit bidirectional transcription, in that they express both sense/mRNAs as well as antisense lncRNAs, appeared to be tumour suppressor genes involved in cell cycle regulation (reviewed in12). More interesting is the observation that those genes exhibiting bidirectional characteristics, such as tumour suppressor genes, are often times found to be epigenetically silenced in human cancers12. From these limited studies as well as a recent studies8,9 much has been gleamed regarding bidirectional genes and discordant regulation of gene transcription.
Mechanistically, this process whereby antisense lncRNAs direct epigenetic silencing complexes to targeted loci in trans appears to involve the lncRNAs interacting with epigenetic protein remodelling complexes and directing them to particular loci in the genome, essentially acting as ‘epigenetic messengers’ (Figure 1). The result of this form of RNA-based transcriptional targeting is stable epigenetic gene silencing (Figure 1b), which has been observed, at least in the case of small RNA guides, to be heritable and passed on to daughter cells13. To date, this mechanism of action has been observed and described with several different genes including p21, p15, PTEN, BDNF, and the iPS related gene OCT-46-9,14.
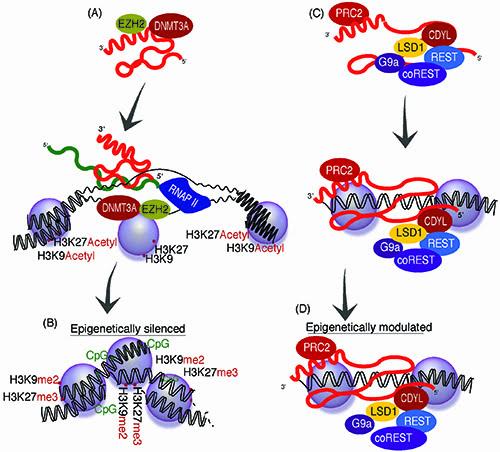

Figure 1: LncRNA epigenetic regulatory pathways. Long non-coding RNAs are expressed from many loci in the genome with some that can function in trans to epigenetically regulate gene expression. (A) Some antisense lncRNAs can interact with those proteins involved in chromatin modification such as DNMT3a, EZH29 while other lncRNAs (B) interact with biomodal controlling PRC2 and LSD1 associated complexes15,18, to epigenetically target particular loci in the genome.
Some lncRNAs that are not necessarily antisense to their protein-coding counterpart have been found to modulate epigenetic states and chromatin architecture. One interesting example of lncRNA function is the bimodal regulating capabilities of HOTAIR, which is derived from the HOXC locus and involved in regulating HOXD in trans15. Similar to another lncRNA, MALAT1, HOTAIR has been observed to be involved in cancer metastasis16,17. Interestingly, HOTAIR has been found to act as a scaffold for the assembly of histone modification complexes18 that can control differential epigenetic states at the HOXD locus, severing to recruit either activating or suppressive epigenetic marks (Figure 1c – 1d). It is however unclear to what extent sequence complementarity is required for HOTAIR targeting to the HOXD locus, unlike antisense lncRNA targeting that involves sequence complementarity in the targeting of epigenetic complexes to protein coding loci and active transcription at the target locus19,20.
There are growing numbers of examples of lncRNA functions involving gene regulation with varying mechanistic themes that for the most part do not rely on epigenetic remodelling of the targeted locus21,16,18,22. Examples abound ranging from Neat123,24, a component of paraspeckles, MALAT-1, a lncRNA involved in subnuclear structure and possibly splicesome function25,25, and Gomafu, a lncRNA involved in schizophrenia26. Clearly, the varied complexities of lncRNA function and regulatory capabilities are only just now becoming apparent. What is noteworthy is the notion that some lncRNAs are active in regulating particular genes by modulating the local epigenetic environment through epigenetic silencing, a mechanism of action that has been shown to have long lasting and heritable potential27. Such a mechanism of action suggests a deep mechanistic of action that has the potential to potently control gene expression.
Mimicking lncRNA pathways with small RNAs: suppressing or activating genes
Around the time pervasive transcription was being realised in the mouse5, it had been observed that small interfering RNAs (siRNAs) could control gene transcription by targeting epigenetic silencing complexes to homology containing loci in human cells28. A year later it was determined that mechanistically only the small antisense RNA (sasRNA) of the siRNAs and DNA methyltransferase 3a (DNMT3a) was required for the observed RNA directed transcriptional gene silencing (TGS)29. Mechanistically, we now know that the sasRNAs interact with a low-copy promoter associated transcript19,20 and recruit epigenetic silencing complexes consisting of DNMT3a29-31, Enhancer of Zeste 2 (EZH2)13, Histone deacetylase 1 (HDAC-1)30 and Argonatues (AGO)32,33 (Figure 2a – 2c) to homology containing loci. This mechanism of RNA directed TGS has now been used to target the stable epigenetic silencing of several genes involved in cancer6,20,34 and diseases such as HIV35-39.
The discovery that sasRNAs can direct TGS in human cells spurred an investigation into the ability to target different gene promoters. During one such investigation, it was observed that promoter targeting, specifically to Adenosine and Thymine rich regions in promoter, resulted in transcriptional activation of the targeted gene40-42. Later studies by a different group on the p21 gene, which was observed previously to be activated by siRNAs targeted to the p21 promoter40, indicated that the mechanism of activation was the result of the siRNAs targeting an antisense long ncRNA6. Such a result, of siRNAs targeting the antisense counterpart, are possible because unlike sasRNAs, siRNAs possess two strands, which have been observed to target both strands of a particular locus, e.g. either sense or antisense transcripts if they are present for a particular targeted mRNA43.
These fortuitous observations of siRNAs targeting an endogenous regulatory lncRNA resulting ultimately in gene activation were some of the first in human cells to indicate that long antisense ncRNAs are functional endogenous modulators of gene expression and epigenetic states (reviewed in44). Such a notion presents the intriguing idea that with some level of knowledge of a particular genes endogenous regulatory lncRNA network, one can target the activation of particular genes expression. This targeted gene activation is done by suppressing the endogenous gene regulatory lncRNA, which leads ultimately to de-repression of lncRNA mediated regulation (Figure 2d – 2f). This process is intriguing as it suggests that the mechanism of lncRNA action (Figure 1a and 1b) can be perturbed in a targeted manner using either RNA interference or antisense oligonucleotide targeting of the lncRNA complex (Figure 2d – 2f respectively); resulting in a loss of lncRNA function and ultimately transcriptional activation of gene expression (Figure 2f). Alternatively, if the promoters for particular regulatory lncRNAs are known one can design sasRNAs to target TGS (Figure 2a – 2c) to the lncRNA promoter34. The result of this targeting can be long-term stable epigenetic silencing of the lncRNA and subsequent stable de-repression (up-regulation) of the lncRNA targeted gene, e.g. TGS of the endogenous lncRNA results in a stable loss of the lncRNA repressor at its targeted gene and ultimately gene activation (Figures 2d – 2f). Indeed, both stable long-term TGS13 and lncRNA promoter targeting34 have been observed previously suggesting this is a plausible methodology to stably activating a particular genes expression.
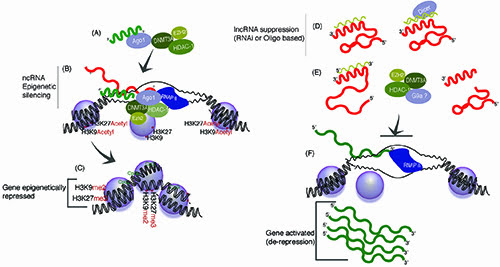

Figure 2: Mimicking and perturbing lncRNA pathways with small regulatory RNAs. (A) Exogenously introduced sasRNAs can target epigenetic silencing to homology containing targeted loci28,29,33, a pathway that miRNAs may also utilise in the nucleus47. (B) This form of targeting requires DNMT3a, HDAC-1, EZH2 and AGO and transcription at the targeted locus (reviewed in 35), and (C) ultimately results in stable epigenetic silencing. (D-E) Suppression of endogenous lncRNA targeted epigenetic silencing can lead to de-repression/activation of the lncRNAs targeted genes expression (reviewed in 44). Regulatory lncRNAs can be targeted with (D) antisense oligonucleotides or RNAi, resulting in (E) conformational changes or degredation of the lncRNA and (F) ultimately de-repression of the lncRNA targeted locus and gene activation.
Collectively, the observations over the last decade portend a new paradigm where it is now possible to specifically modulate a genes expression by mimicking or perturbing endogenous ncRNA directed epigenetic pathways to either turn a gene down or up in a relatively long-term manner. While such a realisation offers an entire new therapeutic reality it is not without its limitations and pitfalls.
Conclusion

One such example is presented here whereby an appreciation for one molecular mechanism, ncRNA direct epigenetic silencing11,35, juxtaposed with current antisense technologies46, suggests it is now possible to modulate stable gene silencing of a targeted locus. The key to this molecular targeting approach is knowledge of the targeted genes functions; genes in this case can be both protein-coding and/or non-coding. Thus targeting depending on the target it is now possible to molecularly control a gene by specifically activating or suppressing its expression. Such RNA directed epigenetic targeting does however also have the potential to stable modulate a genes expression13, suggesting that perturbing lncRNA pathways in this manner can result in long-term heritable epigenetic changes. Indeed such transgenerational epigenetic heritable traits have been observed in model organisms as well as humans (reviewed in11) and may be a key aspect in cellular evolutionary process, suggesting that such superlative control over gene expression may also come at the cost of altering the evolutionary trajectories of the treated cell. While an intriguing notion much remains to be determined with regards to ncRNA mediated epigenetic regulatory networks, as we are only now, just, seeing the proverbial non-coding RNA light.
Acknowledgements: The project was supported by the National Institute of Allergy and Infectious Disease (NIAID) P01 AI099783-01 to KVM and the National Cancer Institute (NCI) R01 CA151574, National Institutes of Health (NIH) R01 CA153124 to KVM and Peter K Vogt.
Biography


References
- Rosenbloom, K.R. et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 40, D912-7 (2012)
- Banfai, B. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22, 1646-57 (2012)
- Malecova, B. & Morris, K.V. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther 12, 214-22 (2010)
- Turner, A.M. & Morris, K.V. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques 48, ix-xvi (2010)
- Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564-6 (2005)
- Morris, K.V., Santoso, S., Turner, A.M., Pastori, C. & Hawkins, P.G. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4, e1000258 (2008)
- Yu, W. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202-6 (2008)
- Hawkins, P.G. & Morris, K.V. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcr 1, 165-175 (2010)
- Johnsson, P. et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20, 440-6 (2013)
- Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559-63 (2005)
- Morris, K.V. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 4 (2009)
- Morris, K.V. & Vogt, P.K. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle 9 (2010)
- Hawkins, P.G., Santoso, S., Adams, C., Anest, V. & Morris, K.V. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res (2009)
- Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol (2012)
- Rinn, J.L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311-23 (2007)
- Gupta, R.A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071-6 (2010)
- Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature (2013)
- Tsai, M.C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689-93 (2010)
- Han, J., Kim, D. & Morris, K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A 104, 12422-7 (2007)
- Napoli, S., Pastori, C., Magistri, M., Carbone, G.M. & Catapano, C.V. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J (2009)
- Vastenhouw, N.L. et al. Gene expression: long-term gene silencing by RNAi. Nature 442, 882 (2006)
- Kretz, M. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231-5 (2013)
- Fox, A.H., Bond, C.S. & Lamond, A.I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16, 5304-15 (2005)
- Mao, Y.S., Sunwoo, H., Zhang, B. & Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13, 95-101 (2011)
- Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39, 925-38 (2010)
- Barry, G. et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry (2013)
- Vassoler, F.M. & Sadri-Vakili, G. Mechanisms of transgenerational inheritance of addictive-like behaviors. Neuroscience (2013)
- Morris, K.V., Chan, S.W., Jacobsen, S.E. & Looney, D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289-92 (2004)
- Weinberg, M.S. et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12, 256-62 (2006)
- Turner, A.M., De La Cruz, J. & Morris, K.V. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Mol Ther 17, 360-8 (2009)
- Ross, J.P., Suetake, I., Tajima, S. & Molloy, P.L. Recombinant mammalian DNA methyltransferase activity on model transcriptional gene silencing short RNA-DNA heteroduplex substrates. Biochem J 432, 323-32 (2010)
- Kim, D.H., Villeneuve, L.M., Morris, K.V. & Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13, 793-7 (2006)
- Janowski, B.A. et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol (2006)
- Ackley, A. et al. An Algorithm for Generating Small RNAs Capable of Epigenetically Modulating Transcriptional Gene Silencing and Activation in Human Cells. Mol Ther Nucleic Acids 2, e104 (2013)
- Morris, K.V. RNA-Directed Transcriptional Gene Silencing and Activation in Human Cells. Oligonucleotides (2009)
- Suzuki, K., T. Shijuuku, T. Fukamachi, J. Zaunders, G. Guillemin, D. Cooper, and A. Kelleher. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. Journal of RNAi and Gene Silencing 1, 66-78 (2005)
- Suzuki, K. et al. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem (2008)
- Turner, A.M., Ackley, A.M., Matrone, M.A. & Morris, K.V. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum Gene Ther 23, 473-83 (2012)
- Lim, H.G., Suzuki, K., Cooper, D.A. & Kelleher, A.D. Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro. Mol Ther 16, 565-70 (2008)
- Li, L.C. et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 103, 17337-42 (2006)
- Place, R.F., Li, L.C., Pookot, D., Noonan, E.J. & Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105, 1608-13 (2008)
- Janowski, B.A. et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol (2007)
- Wei, J.X. et al. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS One 4, e5382 (2009)
- Weinberg, M.S. & Morris, K.V. Long non-coding RNA targeting and transcriptional de-repression. Nucleic Acid Ther 23, 9-14 (2013)
- Clark, M.B. et al. The reality of pervasive transcription. PLoS Biol 9, e1000625 (2011)
- Kole, R., Krainer, A.R. & Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11, 125-40 (2012)
- Kim, D.H., Saetrom, P., Snove, O., Jr. & Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A 105, 16230-5 (2008)
Related topics
Genomics, RNAs, Therapeutics


