Cancer drug screening: the historical perspective
Posted: 20 July 2018 | Alexander Sim - AMS Biotechnology Ltd, Dr Sumeer Dhar (AMS Biotechnology Ltd), Graham Robertson (University of Strathclyde), Michele Zagnoni - University of Strathclyde, Theresa Mulholland (University of Strathclyde) | No comments yet
Chemotherapy with cytotoxic and growth inhibitory drugs have played an important role in cancer therapy, used either alone or in combination with other treatment modalities such as surgery, radiation or biological therapy. Chemotherapy, in most instances, was the only alternative treatment for metastatic cancer – mainly given as drug combinations…

Conventional chemotherapy has shown to produce better response rates for many tumours resulting in the cure of a considerable percentage of these patients. However, the chemotherapeutic treatment has proved to be poorly effective, especially for patients presented in advanced stage of cancer. The first documented use of clinical chemotherapy was observed in 1942, when the alkylating agent nitrogen mustard (belonging to the class of drugs acting through alkylation of DNA) was used to obtain brief clinical remission in a patient with lymphoma.1 Subsequently, various reports indicated the activity in Hodgkin´s disease, lymphoma and chronic lymphocytic and myelocytic leukaemia. With increasing knowledge of basic cellular biochemistry, another class of antitumour agents, the antimetabolites, were discovered.2
Subsequently, additional classes of cytotoxic drugs (eg, anthracyclines, primarily inhibitors of enzyme Topoisomerase II) were discovered,3 followed by the development of the closely related analogue doxorubicin, which was found to have superior activity compared with daunorubicin. Doxorubicin was observed to have a broad-spectrum activity against leukaemias, lymphomas and solid tumours.4 Vinca alkaloids were added to the therapeutic arsenal with their discovery in the late 1960s. These drugs were found to be effective in inducing tumour response in many malignancies and used as standard of care treatment of acute leukaemias, lymphoma and small cell lung cancer. However, development of cisplatinum in the late 1970s ushered in a new class of anticancer drugs with a broad range of antitumour activity. Cisplatinum was initially observed to induce high cure rates in testicular carcinoma and was established as the standard of care for many cancer types, such as small cell lung cancer, ovarian carcinoma, bladder cancer, cervical and head and neck cancer.5,6 Following these, the repertoire of standard of care drugs has continuously been updated with discoveries of taxanes (paclitaxel, docetaxel) and antimetabolites (including Fludarabin, 2-Chlorodeoxyadenosine (CdA)), which were shown to have activity against leukaemias and non-Hodgkin’s lymphomas. Many of these cytotoxic drugs have been given either as single or pairwise, but also as triple combinations, depending on the stage of the disease.
Anticancer drug discovery has continuously evolved over the past several decades from reliance on empirical cell-based screening to studying antiproliferative activities and moving on to more target-based approaches, with the aim of identifying a specific target / culprit that a drug could target to achieve tumour cell killing, leaving healthy cells intact. Researchers at NCI, US, working on chronic myelocytic leukaemia (CML), observed changes in one of the chromosomes – later termed Philadelphia chromosome – present as a fusion gene (known as BCR-ABL). Owen Witte and his colleagues at the University of California, LA, US, later proved that the formation of BRL-ABL in blood cells caused CML.7, 8
During the search for a drug that could be used for curing patients with this abnormality in the chromosome, Brian Drucker, Oregon Health and Science Center, and his colleague identified a drug that was incredibly good at killing CML cells. The drug –named STI-571 and later renamed imatinib (Gleevec) – blocked the activity of the BCR-ABL fusion protein thereby resulting in remission in 98% of patients over the five-year period. This approach to treating cancer patients with a specific targeted therapy revolutionised the way cancer therapy was handled, which previously had operated on a ‘one size fits all’ philosophy. Since then, many targeted drugs have been approved for treatment of various cancers and so too have the methods for discovery and development of new anticancer agents, which have included targeted molecules, monoclonal antibodies and T cell-based therapeutic approaches (26 approved immuno-oncology agents).9 Yet, some specific cancer types, such as glioma and pancreatic, remain intractable to new therapies and advanced cancers represent critical areas of unmet therapeutic need. High attrition rates during clinical development of new cancer therapies persist, despite investments made in the R&D for discovery and development of new molecules for therapy. Failure to translate promising preclinical drug candidates into clinical success exposes the limitations of our current drug discovery models to predict clinical outcomes.
For many approved cancer drugs, the patient’s benefit is often initially small and incremental improvement in treatment efficacy and duration may take many years of experimental medicine studies within clinical trials. The challenges in developing successful cancer therapeutics are not just limited to the discovery of new drug therapies, but also the availability of robust preclinical ex vivo models. Moreover, traditional drug discovery and discovery medicine activities have relied upon rudimentary 2D cell monolayer models, 3D clonogenic assay platforms and small animal models, mostly utilising established cancer cell lines. It is now apparent that such preclinical models do not accurately recapitulate the complex pathophysiology of cancers found in patients and poorly predict clinical response. Therefore, development of more predictive, patient-specific models of human cancer are pivotal in the quest for profiling of novel anticancer drugs, either as single entities or as drug combinations. Oncologists recognised several decades ago the biological similarities between cells grown ex vivo in 3D culture systems and avascular tumours in vivo.10 The better the in-vitro models reflect the function and structure of their in vivo counterparts, the more predictive the cell-based assays become. Currently, the use of ex vivo 3D assay platforms is being pursued by researchers as a tool for drug activity response and monitoring potential changes at subcellular levels to identify downstream targets post drug treatment.
Additionally, these platforms are extensively being used as a drug development tool, as it has been shown to closely simulate the tumour microenvironment within the in vitro milieu.11,12,13 It has previously been reported that the drug response of cancer cells is not just determined by the inherent characteristics of epithelial tumour cells, but is also controlled by signals derived from other cells (stromal / fibroblasts / distinct immune cell compartments) within the tumour microenvironment.14,15,16, 17
It has been reported that the abnormal vascularistion of solid tumours leads to the generation of tumour microenvironments that are chronically starved of oxygen and nutrients.18,19
As a result, cells residing in such an environment demonstrate altered phenotypic characteristics when compared with the cells located in more vascularised regions around the outer core.20,21 Therefore, there has been an immense drive towards developing methods that can exploit the altered phenotype of tumours. One of the earliest 3D-related effects, which were correlated to in vivo observations, included multicellular drug resistance (MDR). Cancer cells grown as 3D spheroids generally display reduced drug sensitivity compared with 2D monolayer cultures.22 These observations indicate that the use of 3D spherical microtissues may enable improved discrimination of the most active anti-cancer drugs with improved therapeutic index.
State of current 3D ex vivo platforms and their utility
Recent technological advances in high-throughput and content phenotypic screening and 3D multicellular assay methods, have opened doors for reshaping several key processes during drug discovery and development. The emergence of technologies such as 3D perfusion systems and 3D microfluidics formats, with special focus on multicellular tumour spheroids (MCTS) systems, present robust platforms to conduct drug screening and early development programmes in cost effective and efficient ways, using clinically relevant cell types derived from multiple cancers. Within the context of 3D ex vivo platforms, microfluidic technologies have emerged as potentially highly relevant ex vivo tools in cancer research.23,24,25 This rapidly expanding technology relies on the use of small channels equipped for handling small fluids and, therefore, small amounts of chemicals and cell material. These platforms have shown to represent physiological relevance over other in vitro technologies, allowing precise control of the cellular, physical and biochemical environment; and the ability to study cancer-immune interactions, making them a complementary platform for preclinical in vivo models.26,27
AMS Biotechnology and its collaborators from Strathclyde University in Glasgow, has established a unique 3D ex vivo microfluidics platform to enhance cancer drug screening and early development process, aiming to transform the poor success rates of translating preclinical drug discovery and to accelerate experimental medicine programmes towards clinical success. This platform offers great promise to create cancer models utilising immortalised standard cell lines, panels of CRISPR cell lines with known clinically relevant mutations, cells derived from xenograft models, as well as patient-derived primary cells. The technology provides a versatile 3D ex vivo system to assess efficacy of small and large molecules in pairwise and radio-chemo combinations.
AMSBIO-Oncoscreen 3D ex vivo microfluidics platform
The microfluidics system developed consists of a miniaturised and high-throughput 3D ex vivo assay platform, which provides a robust, customisable and cost-effective screening system (Mullholland et al, publication under review). The novel characteristics of this platform enable rapid decision making to prioritise the most promising drug candidates, biomarkers and drug combination strategies for preclinical drug discovery and development. An advantage of the platform is the ability of cells to form spheroids without the presence of exogenously added matrices / scaffolds to help cells polarise into formation of spheroids, as the endogenously produced ECMs by cells help polarisation and induce cell contact. This, together with precise fluid control, enables formation of compact spheroids in less than five days, especially cells derived from patient biopsies or PDXs. Therefore, the system enables long-term culture as well as fractionated chemo- and targeted-therapy to study drug efficacy, as well as acquired drug resistance.
The platform has been validated by assessing the efficacy of standard of care compounds against multiple cancer types including glioblastoma, prostate, breast, lung and pancreatic cancer. This platform offers the ability to perform multiparametric end point measurements that include (but are not limited to) viability measurement, changes in spheroid size and shape, and assessing the temporal evolution of spheroid response post drug treatment. The system also allows researchers to retrieve spheroids for proteomics and transcriptomics analysis, as well as changes in the spheroid biomarker status using immunohistochemistry techniques (Figure 1).
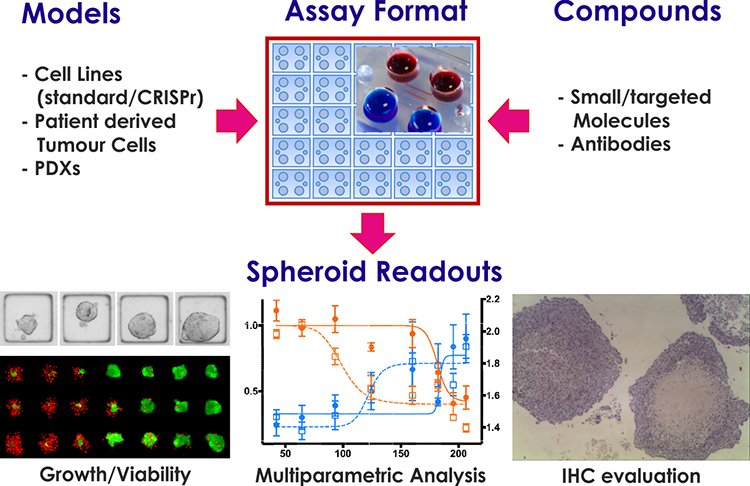

Figure 1: AMSBIO 3D ex vivo cancer drug screening platform.
In conclusion, among various 3D ex vivo platforms available, AMSBIO 3D ex vivo microfluidics platform provides a versatile tool well suited for cancer drug discovery, early development and profiling of broad spectrum of molecules including small and targeted molecules.
Biographies










References
- Calbresi P, Parks RJ (1975). Alkylating agents, antimetabolites, hormones and other antiproliferative agents. IN: Goodman and A. Gilman (ed.), The Pharmacological Bases of Therapeutics., pp. 1254-1268, Macmillan, New York.
- Pratt W, Ruddon R (1979). The Anticancer Drugs, Oxford University Press, New York, pp 98-122.
- Pigram W, Fuller W, Hamilton L (1972). Stereochemistry intercalation: interaction of daunorubicin with DNA. Nature New. Biol., 235, 123-125.
- De Vita Jr VT (1997). Principles of cancer management. Chemotherapy. In: V:T. DeVita Jr, S. Hellman and SA Rsenberg (eds), Principles and Practice if Oncology. Vol. 1, pp 333-347. Lippincott-Raven, Philadelphia, PA.
- Loehrer PJ, Einhorn LH (1984). Cisplatinum. of Intern. Med. 100, 704-713.
- O´Dwyer P, Johnson S, Hamilton T (1997). Cisplatimun and its analogs. In: V. De Vita Jr, S. Hellman and S Rosenberg (eds), Cancer, Principles & Practice of Oncology, 418-432, Lippincott-Raven, Philadelphia.
- Wong S, Witte ON (2004). The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 22:247-306.
- Witte O (2001). The role of Bcr-Abl in chronic myeloid leukemia and stem cell biology. Semin Hematol. Jul;38(3 Suppl 8):3-8.
- Tang J, Shalabi A, Hubbard-Lucey VM (2017). Comprehensive analysis of the clinical immuno-oncology landscape. Annals of Oncology. 0, 1-8.
- Hirschhaeuser F, Menne H, Dittfeld C, et al. (2010). Multicellular tumor spheroids: an underestimated tool is catching up again. Jour of Biotechno, 148(1), 3–15.
- Mizushima H, Wang X, Miyamoto S, Mekada E (2009). Integrin signal masks growth promoting activity of HB-EGF in monolayer cell cultures. J Cell Sci. 122:4277-86.
- Kondo J, Endo H, Okuyama H, Ishikawa O. et al. (2011). Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci . Apr 12;108 (15):6235-40.
- Yoshii Y, Furukawa T, Waki A, Okuyama H, Inuoue M, Itoh M et al. (2015). High-throughput screening with nanoimprinting 3D culture for efficient drug development by mimicking the tumor environment. Biomaterials 5:278-89.
- Sun Y, Campisi J, Higano, C. Beer TM. et al. (2012). Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Sep;18 (9):1359-68.
- Morikawa, T. Shee, K. et al. (2012). Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Jul 26;487 (7408):500-4.
- Ostman A (2012). The tumor microenvironment controls drug sensitivity. Sep;18 (9):1332-4.
- Horman SR, To J, Lamb J, Zoll JH, Leonetti N, Tu B, Moran R, Newlin R, Walker J and Orth AP (2017). Functional profiling of microtumors to identify cancer associated fibroblast-derived drug targets. Oncotarget, 59, 99913-99930.
- Harris AL (2002). Hypoxia-a key regulatory factor in tumor growth. Nat. Rev. Cancer 2, 38-47.
- Mellor HR & Callaghan R (2008). Resistance to chemotherapy in cancer: a complex and integrated cellular response. Pharmacology, 81, 275-300.
- Sahin AA, R JY, el-Naggar AK, Wilson PL et al. (1991). Tumor proliferative fraction in solid malignant neoplasms. A comparative study of ki67 immunostaining and flow cytometric determination. Am J Clin Pathol. 96, 512-519.
- Gardener L.B. et al. (2001). Hypoxia inhibitsG1/S transition through regulation of p27expression. J. Biol. Chem. 276, 7919-7926.
- Desoize B, Jardillier J. (2000). Multicellular resistance: a paradigm for clinical resistance? Cri Rev Oncol Hematol, Nov-Dec; 36 (2-3): 193-197.
- Terrell-Hall TB, Nounou MI, El-Amrawy F, Griffith JIG, Lockman PR. (2017). Trastuzumab distribution in an in-vivo and in-vitro model of brain metastases of breast cancer. Oncotarget. Jul 26;8(48): 83734-83744.
- Lanz HL, Saleh A, Kramer B, Cairns J, Ng CP, Yu J, Trietsch SJ, Hankemeier T, Joore J, Vulto P, Weinshilboum R, Wang L. (2017). Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer. Nov 2;17(1):709.
- Yin X, Mead, E.B, Safaee H, Langer R, Karp JM. (2016). Engineering Stem Cell Organoids. Cell Stem Cell, January 7 (18): 25-38.
- Boussomier-Calleja A, Li R, Chen MB, Wong SC and Kamm RD. (2016). Microfluidics: A new tool for modelling cancer immune interactions. Trends Cancer. January 1;2(1): 6-19.
- Jenkins RW, Aref AR, Lizotte PH, Ivanova E, et al.,(2017). Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 8(2):196-215.
The rest of this content is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- quarterly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Related topics
chemotherapy, Drug Targets, Oncology, Radiotherapy, Research & Development, Screening, Therapeutics


