Finding a cure for neglected tropical diseases with cell-based assays
Posted: 14 February 2016 | David Shum (Institut Pasteur Korea) | No comments yet
Neglected tropical diseases pose a significant threat to public health and affect nearly one billion people in tropical and sub-tropical regions throughout the world. There is a pressing need to identify new therapeutics for NTDs and many developed nations are participating in drug discovery efforts…

Moreover, the rise of drug resistance towards once effective treatments has become a serious concern and new improved methods for discovery are needed. Cell-based assay systems that accurately reflect the NTD’s pathogenesis are currently in development for high-throughput screening (HTS). Utilising a phenotypic approach, chemical libraries are rapidly tested for active compounds against the pathogen without detriment to the cell. In parallel, the host-pathogen interactions are being interrogated through the use of functional genomics to gain a better understanding of fundamental biological processes involved in disease progression. In this contribution, we review several cell-based assays for NTD research and discuss their importance during the early stages of the drug discovery pipeline.
Introduction


NTDs are represented by a diverse group of 17 infectious pathogens that include viruses (two), bacteria (four) and parasites (eight parasitic helminths and three parasitic protozoans)2. These infectious diseases are endemic in poorer and developing regions of Asia, Africa, and the Americas where access to basic healthcare remains a burden. Additionally, NTDs are characterised by their lack of public attention and research funding, and often there are no specific treatments available. Although largely confined to certain regions, these diseases have the potential to spread throughout the rest of the world due to air travel and globalisation. The socioeconomic impacts can be felt with costs of billions in dollars every year, highlighting the need for the development of new therapeutic treatments and as a result, the World Health Organization (WHO) has targeted several NTDs for eradication by 2020.
NTD pathogens utilise different strategies to infect, replicate and survive, creating a challenge for therapeutic interventions3. NTDs arising from parasites display complicated life cycle stages and involve multiple developmental forms in the vector and host. NTDs triggered by viruses are transmitted through infected vectors and then orchestrate numerous biological processes in the host to replicate. Bacterial NTDs are spread by host contact and characterised by several stages of infection involving the skin, bones and cartilage. Subsequently, many complex interactions play a key role in disease progression and current treatments may eventually prove inadequate as drug-resistant strains have already emerged.
Advances in technology have resulted in new methods that enable rapid testing in the context of biologically relevant disease models. Cell-based assays utilising phenotypic approaches that accurately reflect the disease’s pathogenesis through various stages within the host are being developed. Several of these methods have already been in use for HTS and are of critical importance in the early drug discovery phase. In this contribution, we focus on cell-based assays in use for research on parasitic protozoan and viral NTDs, which offer promise for new therapeutic interventions.
Phenotypic approach to NTDs
Given the pressing need to rapidly develop new therapeutics for NTDs, cell-based assays are ideally suited and in combination with automated microscopy are a powerful approach in the early drug discovery pipeline. Cell-based assays utilise human or murine established lines as surrogates for pathogens to infect, replicate and egress that better mimic the disease’s progression. In these biologically-relevant disease models, infectious pathogens can be tracked by phenotypic readout using various reporter constructs or fluorescent dyes for the visualisation of biological processes involved in their life cycles. By adapting this approach to an HTS environment, a myriad of compounds or RNA interference (RNAi) tools can be screened at a given time to simultaneously evaluate efficacy, permeability, targeting and toxicity. Subtle changes in phenotype can be acquired through imaging and analysed using sophisticated algorithm tools to assess the pathogen’s localisation, quantity and effect.
Cell-based assays are now routinely utilised in HTS mode for primary screening campaigns against large chemical collections (Figure 1). In this screening environment, the assay provides relevant data on compound efficacy, permeability, solubility and cellular toxicity that can used to assess its potential as a drug. Compounds identified to act on the pathogen typically do not have any targeting information. Despite this, phenotypic screening more accurately reflects the in vivo condition of the compound’s effect on a disease model and follow-up techniques have improved allowing for target identification. In an alternative strategy, chemical libraries of known pharmacological properties are screened and compounds can be repurposed for use without the need for additional deconvolution. This effective strategy has been in use for malaria and is now actively being applied to research in NTDs4.
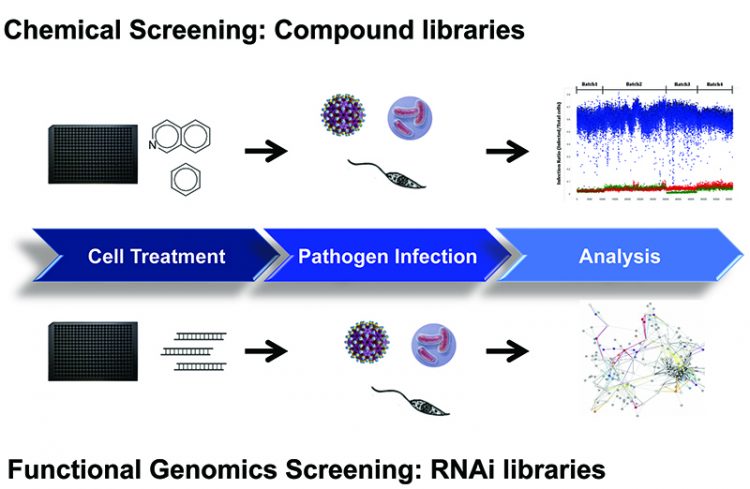

Figure 1: Cell-based assay workflow for chemical and functional genomics screening.
Cell-based assays that are readily used in HTS mode for compound screening are adaptable for RNAi technology using siRNA and shRNA. Systematic knockdown of the gene is performed to identify targets and dissect pathways essential for host-pathogen interactions. The key steps from diagnostics, treatment and eradication is to expand the repertoire of drugs that would target the different mechanisms of the pathogen’s life cycles. These strategies have been utilised in recent years for influenza5 and other infectious diseases such as HIV to provide a better understanding of the biology of pathogenesis6. Additionally, cell-based assays have been developed in various stages of pathogen’s life cycle including those involving zoonotic hosts and free-living hosts to provide a comprehensive overview of the disease7. Phenotypic approaches in screening have proven efficient in evaluating compounds, identifying new targets and discovering novel mechanisms, and consequently make an invaluable tool for NTD research.
NTD research

The protozoan parasite Leishmania is characterised by the presence of a unique organelle called kinetoplast and responsible for Leishmaniasis. Leishmaniasis is a disease that primarily affects Latin America, Africa and the Indian subcontinent and is divided into the following types: cutaneous, mucocutaneous and visceral, the latter being the most serious leading as it does to death. Leishmania exist in several forms throughout their life cycle starting as promastigotes in sandflies and intracellular amastigotes after transmission to host. Leishmania assay systems that represent the insect stage have been developed with promastigotes and are easy to handle. However, active compounds typically translate poorly in secondary assays involving host cells.
Cell-based assays for intracellular amastigotes have been developed utilising phenotypic readout to determine inhibition of infection and parasitemia. In this system, the promastigotes are used to infect macrophages and resulting intracellular amastigotes can be detected utilising fluorescent staining dyes (Figure 2A). The nuclei of the cell is first detected and a secondary mask is applied to locate all intracellular amastigotes within the cytoplasmic boundaries. Using an algorithm for analysis, a quantitative measurement can determine the compound’s effect on parasite clearance and the most promising candidates typically have high efficiency without cellular toxicity. This methodology has been used in a screen of 26,500 compounds in human differentiated THP1 macrophages and using a threshold of 55% activity identified 123 candidates8.
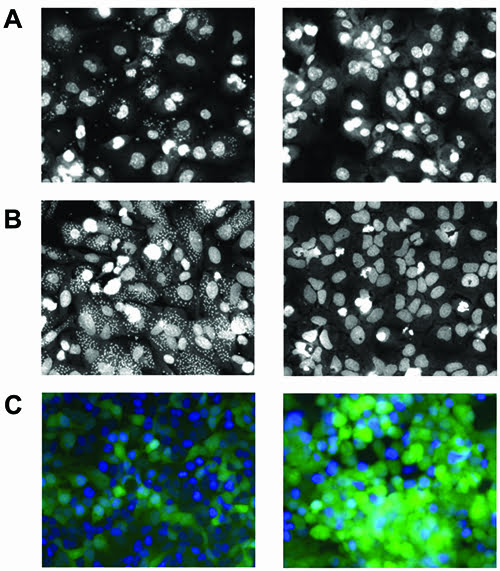

Figure 2A: Leishmaniasis: Differentiated THP1 macrophages with intracellular amastigotes (left: control treatment; right: inhibitor treatment). B: Chagas disease: U2OS cells with intracellular amastigotes (left: control treatment; right: inhibitor treatment). C: Rabies: HeLa cells infected with lyssavirus expressing GFP (left: 18 hour control treatment; right: 36 hour control treatment).
In another study, a genetically modified strain with DsRed2 fluorescent label in mouse macrophages was validated with a small library of known compounds against leishmaniasis9. Here, a fluorescently labelled strain is particularly advantageous allowing for direct visualisation and kinetic studies through pathogenesis. Lastly, a group using the using the same methodology performed an shRNA screen in human U937 macrophage cells to interrogate essential genes involved in host-pathogen interactions10.
Cell-based assay systems using a similar phenotypic model can be readily applied to Chagas disease, which also belongs to the kinetoplast family. Found in Latin America, Chagas disease is caused by protozoan parasite Trypanosoma cruzi and infection results in myocardial abnormalities. Similarly, life cycle stages for T.cruzi starts as trypomastigotes in triatomine bugs and resulting transmission into host form intracellular amastigotes. Cell-based assays have been developed for intracellular amastigotes in different cells since T.cruzi can infect numerous tissues. In a system with U2OS cells, trypomastigotes were infected and resulting intracellular amastigotes were validated against two T.cruzi drugs: benznidazole and nifurtimox11 (Figure 2B). This methodology was applied to rat myoblastic cells in a screen of 20 compounds of known activity12 and in another study with human HUH7 hepatocarcinoma against 909 compounds13.
Rabies is a disease caused by the single-stranded RNA lyssavirus and predominantly found in Africa and Asia. It is transmitted by scratches or bites from an infected animal and results in neurological conditions that ultimately lead to death. At the onset of infection, rabies utilises the host cell to replicate new viruses that attack the central nervous system and further spread to other organs. Traditional methods for antiviral development have relied on cytopathic and plaque reduction assays, which are laborious. Cell-based assays utilising phenotypic readout to evaluate antiviral activity have been developed that monitor both efficacy and cellular toxicity in a rapid manner.
In this system, a recombinant lyssavirus with GFP expression linked to M protein (matrix) was used to infect HeLa cells and their life cycles monitored by live-cell imaging (Figure 2C). Through image analysis, an algorithm identifies the nuclei and green fluorescence intensity within the cell to determine factors that influence early and late stage events. This methodology has been used in a siRNA screen against a focused library of kinase and phosphatase genes at 18 and 36 hour time points. Quantitative changes in green expression are an indication of host-pathogen factors important in viral entry, replication and processing.
Conclusion
As NTD research has been challenging due to underfunding and a perception that these are diseases of poverty, assays that provides critical information on the pathogen in a biologically relevant disease model will accelerate the drug discovery process. Moreover, therapeutic treatments may also become obsolete as drug-resistant strains emerge, and the development of drugs with new modes of actions is needed. Cell-based assays in which the pathogens are used to infect human or murine established lines are currently in development and utilise phenotypic readout for detection and analysis. Once adapted to microtiter plate format, the cell-based assays can eventually be implemented into the HTS pipeline against compound and RNAi libraries. These systems have already been tested against leishmaniasis, Chagas disease and rabies in which large chemical libraries were screened and targets have been interrogated. Given the current status and needs of these diseases, cell-based assays are a reliable model for understanding pathogenesis and an efficient tool towards the development of new therapeutic strategies.
Biography


References
- Mackey TK, Liang BA, Cuomo R, Hafen R, Brouwer KC, Lee DE. Emerging and reemerging neglected tropical diseases: a review of key characteristics, risk factors, and the policy and innovation environment. Clin Microbiol Rev. 2014; 27(4), 949-979
- Buscaglia CA, Kissinger JC, Aguero F. Neglected Tropical Diseases in the Post-Genomic Era. Trends Genet. 2015; 31(10), 539-555
- http://www.who.int/neglected_diseases/diseases/en/
- Nzila A, Ma Z, Chibale K. Drug repositioning in the treatment of malaria and TB. Future Med Chem. 2011; 3(11):1413-1426
- Atkins C, Evans CW, Nordin B, Patricelli MP, Reynolds R, Wennerberg K, Noah JW. Global Human-Kinase Screening Identifies Therapeutic Host Targets against Influenza. J Biomol Screen. 2014; 19(6):936-946
- Jiang WM, Zhang XY, Zhang YZ, Liu L, Lu HZ. A high throughput RNAi screen reveals determinants of HIV-1 activity in host kinases. Int J Clin Exp Pathol. 2014; 15;7(5):2229-2237
- Paveley RA, Mansour NR, Hallyburton I, Bleicher LS, Benn AE, Mikic I, Guidi A, Gilbert IH, Hopkins AL, Bickle QD. Whole organism high-content screening by label-free, image-based Bayesian classification for parasitic diseases. PLoS Negl Trop Dis. 2012; 6(7):e1762
- Siqueira-Neto JL, Moon S, Jang J, Yang G, Lee C, Moon HK, Chatelain E, Genovesio A, Cechetto J, Freitas-Junior LH. An image-based high-content screening assay for compounds targeting intracellular Leishmania donovani amastigotes in human macrophages. PLoS Negl Trop Dis. 2012; 6(6):e1671
- Aulner N, Danckaert A, Rouault-Hardoin E, Desrivot J, Helynck O, Commere PH, Munier-Lehmann H, Späth GF, Shorte SL, Milon G, Prina E. High content analysis of primary macrophages hosting proliferating Leishmania amastigotes: application to anti-leishmanial drug discovery. PLoS Negl Trop Dis. 2013; 7(4):e2154
- Ovalle-Bracho C, Londono-Barbosa DA, Franco-Munoz C, Clavijo-Ramirez C. Functional evaluation of gene silencing on macrophages derived from U937 cells using interference RNA (shRNA) in a model of macrophages infected with Leishmania (Viannia) braziliensis. Parasitology. 2015; 142(14), 1682-1692
- Moon S, Siqueira-Neto JL, Moraes CB, Yang G, Kang M, Freitas-Junior LH, Hansen MA. An image-based algorithm for precise and accurate high throughput assessment of drug activity against the human parasite Trypanosoma cruzi. PLoS One. 2014; 9(2):e87188
- Alonso-Padilla J, Cotillo I, Presa JL, Cantizani J, Peña I, Bardera AI, Martín JJ, Rodriguez A. Automated high-content assay for compounds selectively toxic to Trypanosoma cruzi in a myoblastic cell line. PLoS Negl Trop Dis. 2015; 9(1):e0003493
- Engel JC, Ang KK, Chen S, Arkin MR, McKerrow JH, Doyle PS. Image-based high-throughput drug screening targeting the intracellular stage of Trypanosoma cruzi, the agent of Chagas’ disease. Antimicrob Agents Chemother. 2010; 54(8):3326-3334
Related topics
Assays, Cell-based assays, Disease research, Drug Discovery, High Throughput Screening (HTS), RNAs, Screening, Therapeutics


