Informatics infrastructure for public-private collaborations in neglected disease research
Posted: 13 December 2017 | Julio Martin | No comments yet
Efforts to develop new medicines for diseases of the developing world (DDW) have been somewhat fragmented in the past and progress has been limited, despite considerable investment. Public-private partnership (PPP) is becoming an essential model for research in neglected disease areas. However, collaboration on this scale presents unique challenges, some of which can be well managed with the right informatics tools…


GlaxoSmithKline (GSK) has pioneered a model of true open partnership with academia through the Tres Cantos Open Lab Foundation, a ground-breaking PPP initiative set up at GSK’s dedicated DDW research facility at Tres Cantos in Madrid. Since its inception in 2011, the Tres Cantos Open Lab Foundation has managed more than 50 projects of drug discovery efforts against tuberculosis, malaria and kinetoplastid diseases (including trypanosomiasis, ie, African sleeping sickness and Chagas disease, and leishmaniasis). It is the world’s first open laboratory to investigate diseases of the developing world.
By combining the resources of GSK with the dedication and focus of external partners, world-class scientists can test ideas and develop new tools and assets working hand-to-hand with GSK scientists on an industrial scale. Infrastructure technologies that make this possible are shared among an internal staff of 150 dedicated scientists and 15-20 concurrent visiting scientists, working in partnership with key stakeholders and institutions in the field (Figure 1). Research data must be secure, but to facilitate productive collaboration it must also be accessible in real time from around the world.
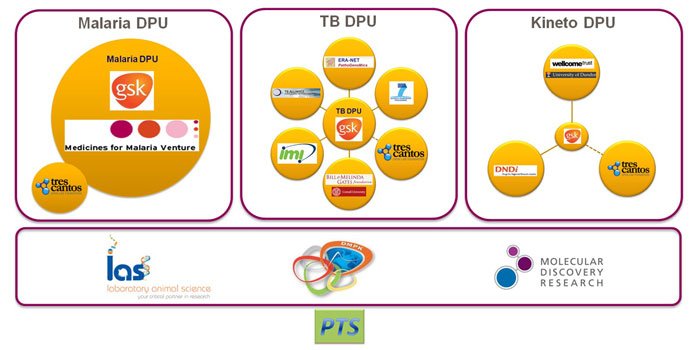

Figure 1: DDW research in GSK has an extensive network of internal and external partnerships and projects. (DPU = Discovery Performance Unit)
The Open Lab Foundation aims to stimulate collaboration by giving external academic partners access to GSK compounds, infrastructure and drug discovery expertise. It hopes to accelerate developments in DDW research areas spanning target discovery and validation, compound screening, and lead identification and optimisation. Critically, the end point is not commercial return, but the development of drugs and the provision of access to healthcare for populations with the greatest need. It is an approach that encompasses free access to GSK resources and know-how (the Open Lab), sharing data and compounds with the global research community (Open Source), and flexibility with IP (Patent Pool).
Optimising data management
Collaboration between GSK and multiple partners on this scale presents significant challenges in relation to data management. All too often, the exchange of information within the research community takes place through PDF, Excel or other flat files, which offer no context, cannot be properly mined, interrogated, or analysed, and end up in data silos as ‘dead data’. The information challenge is compounded when organisations carry out collaborative research, or work with outsourcing partners who need to share, compare and combine disparate data, but remain confident of data security and integrity.
To optimise data management, collaborative consortia need a flexible and user-friendly informatics infrastructure that can underpin data entry, handling and storage, without compromising security. Ease of data input, data query, retrieval and visualisation, as well as optimised data tracking, exchange, integrity and security, are critical on the list of ‘must haves’, as is the ability to manage both biological and chemical data.
GSK’s kinetoplastid programme at the DDW site, as an example, is centred on the discovery of innovative medicines against leishmaniasis, Chagas disease and sleeping sickness. In 2015, GSK made a new set of 592 compounds publicly available as an open resource for drug discovery against…
kinetoplastid diseases.1 The chemical structures and biological annotation are published in open access journals and databases. Likewise, the kinetoplastid chemical box, aka Tres Cantos Anti-Kinetoplastid Screening (TCAKS), is available on request to external collaborators who agree with the ultimate objective of publishing emerging data in the public domain. It is expected that the information disclosed so far, plus that generated by the investigators testing the compounds, will enable the scientific community to address relevant research questions and seed lead discovery programmes that eventually deliver innovative treatments for these important but neglected, diseases.
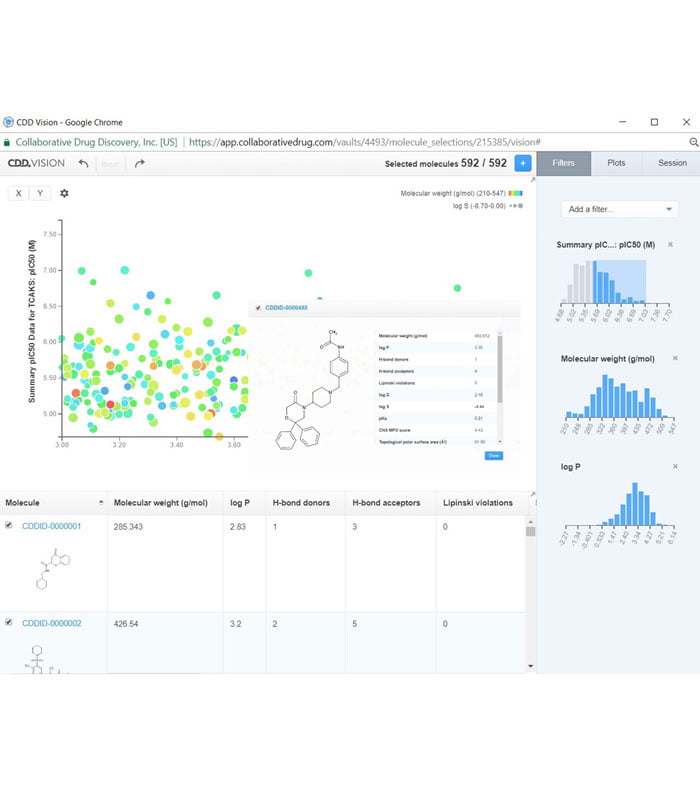

Figure 2: CDD Vault provides a web interface that GSK Kinetoplastid DPU uses to share, manage, and analyse data with its collaborating partners
With this aim, the Kinetoplastid Discovery Performance Unit (DPU) of GSK selected Collaborative Drug Discovery’s CDD Vault (CDDVault.com) as its repository for all the TCAKS-related data generated internally and through its partnerships with external global collaborators. Because this is a hosted, software as a service (SaaS) solution, participating researchers around the world can input, interrogate and mine shared project data and assay results from any web browser (Figure 2). This kind of hosted solution avoids challenging firewall navigation, scales automatically as the projects grow, and saves internal resources for scientific research rather than IT setup, maintenance and upgrades.
The information will be stored for collaborators to explore and analyse from anywhere in the world, secured by two-factor authentication. This will give the global research community new insights into the diseases, their targets and potential drug compounds, regardless of who has made the discoveries. Establishing multiple open partnerships and making data publicly available will hopefully speed drug discovery and development for DDW, reduce failure rate, and leverage new opportunities for collaborative R&D.
Each partnership is structured in a three-tier hierarchy that defines the exposure of resulting data. Initially, assay results and other data are stored and shared bilaterally between only the originator and GSK. Through the second tier of the agreement, the data can be promoted and made accessible to other partners within the community of TCAKS requesters. Finally, when the originator is happy to do so, the data are made public – the third data tier – which is the overarching goal of all the partnerships.
Controlling data sharing
A major benefit is that the CDD Vault allows data sets to be shared exclusively with select individuals, but when approved by the parties involved, vaults of data can be promoted to wider audiences, r even open publication. Furthermore, because it is hosted in the cloud, it removes the need to shuttle data across firewalls, eliminating the chance of inappropriate access to in-house, proprietary research data. The exchange of data between GSK and the partners remains silent and secure until the partners agree that they want to release it.
Critically, this approach allows GSK to maintain all compounds, biological annotations and data from internal and external principal investigators in a single repository. Researchers can interrogate the database retrospectively, not just for their own data, but for results and data submitted and released by other partners, with confidentiality maintained. The initiative’s external collaborating scientists have embraced the concept of sharing their data with other authorised researchers for the benefit of the DDW research community.
Having data available in a single repository also makes it easier to ensure that research is not being repeated by different external collaborators. The Kinetoplastid DPU, as administrator, can mine the CDD Vault to detect overlapping research, even at the proposal stage, and this opens up the potential to put researchers who are working in similar areas in contact with each other.
The goal of GSK with respect to the Kineto Boxes is to make experimental data publicly available and help springboard drug development for DDWs. Publication by external partners is thus a measure of the success of the initiative, which is already starting to bear fruit. Data emerging from the programme are now being presented at conferences by principal investigators, and the first peer-reviewed paper on the analysis of GSK’s kinetoplastid compound set has been published.2 This has been a very encouraging milestone, which will hopefully be followed by many more publications.
The DDW site of GSK and Tres Cantos Open Lab Foundation provide an innovative solution to further the collaborative process of drug discovery for neglected diseases. In addition to supplying the community with research facilities, compound sets, funding, and expertise, they also provide an essential informatics framework that makes storing, analysing, searching, and sharing of critical research results possible. Continued success will drive new collaborations, underpin additional fundraising to further accelerate R&D in the DDW field, and aid the development of effective, safe drugs for some of the world’s most devastating diseases.
Biography
JULIO MARTIN is Director and Head of the Kinetoplastid Discovery Performance Unit (DPU) of GSK R&D at Tres Cantos in Spain. He was previously responsible for ultra-HTS campaigns from screen development to dose-response and preliminary SAR, and was engaged in the development and implementation of new statistical tools and assay technologies for the improvement of HTS efficiency. Dr Martin holds a PhD in Biochemistry from the University of Madrid.
References
- Peña I, Pilar Manzano M, Cantizani J, Kessler A, Alonso-Padilla J, Bardera AI, Alvarez E, Colmenarejo G, Cotillo I, Roquero I, de Dios-Anton F, Barroso V, Rodriguez A, Gray DW, Navarro M, Kumar V, Sherstnev A, Drewry DH, Brown JR, Fiandor JM, Julio Martin J. New Compound Sets Identified from High Throughput Phenotypic Screening Against Three Kinetoplastid Parasites: An Open Resource, Scientific Reports. 2015;5. Article number: 8771. doi:10.1038/srep08771
- Salas-Sarduy E, Landaburu LU, Karpiak JX, Madauss KP, Cazzulo JJ, Agüero F, Alvarez VE. Novel scaffolds for inhibition of Cruzipain identified from high-throughput screening of anti-kinetoplastid chemical boxes, Scientific Reports. 2017;7. Article number: 12073. doi:10.1038/ s41598-017-12170-4
Related topics
Informatics
Related organisations
GlaxoSmithKline


