Browsing for drug targets
Posted: 8 March 2016 | Edward D. Zanders, PharmaGuide Ltd, Cambridge, UK | No comments yet
Here, Edward D. Zanders explains his motivation behind writing ‘Human Drug Targets a Compendium for Pharmaceutical Discovery’ – a collection of current and potential drug targets mapped to human genes curated by the HGNC…


‘Human Drug Targets a Compendium for Pharmaceutical Discovery’ is a collection of current and potential drug targets mapped to human genes curated by the Human Genome Nomenclature Committee (HGNC). This article explains the motivation behind writing the book, some examples of content, and how it is structured to allow the reader to find updated information online.
Thirteen years have now passed since the formal completion of the human genome sequencing project, a period of time sufficient to cover a full drug development cycle from target to marketed drug. The practical value of this project is apparent through the impressive number of innovative medicines that are now in the clinic or in late development that would not otherwise have appeared. The human genome contains approximately 19,000 protein-coding genes and 4600 noncoding RNAs (lncRNAs and microRNAs) (1). Of those that are not targets for existing small molecule or biological drugs, which ones will be the drug targets of the future? There is certainly no shortage of information available to help answer this question. Internet-based tools are indispensible for dealing with the huge amount of available data, but as every web user knows, the results can become overwhelming. One way of dealing with this is to encourage browsing, an activity with which everyone is familiar in many aspects of life and something that has been the subject of academic study in the information sciences, for example by Hjørland (2): “Browsing is a quick examination of the relevance of a number of objects which may or may not lead to a closer examination or acquisition/selection of (some of) these objects. It is a kind of orienting strategy that is formed by our “theories,” expectations and subjectivity.”
Analogue browsing occurs in physical libraries where the reader may seek out a specific book but then become distracted by other volumes within eyesight which may contain more relevant material. Search engine results are the online equivalent, where information is presented in context as a list of hits, some of which may be analogous to the unsought, but useful library book. The ability to browse for information in a convenient book form was one motivation for putting together a compendium of drug targets; it encourages the reader to scan through many human genome entries in the hope that something will catch their interest and perhaps ultimately lead to a significant research investment. Another motivation was to try and capture the speculative comments that many authors make in the discussion section of their biomedical publications, namely “this (phenomenon) could provide a novel therapeutic target” or similar phrases. Just collecting these text strings from the online literature provides a useful list of entries that may contain some genuine opportunities for drug development, while fully recognising that many will just remain speculations.
Layout and organisation
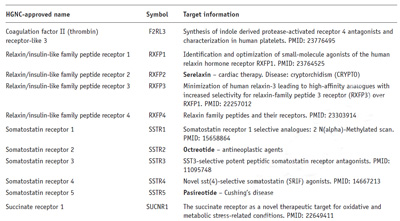
Approximately 9,600 protein and 170 noncoding RNA entries are listed according to the name and symbol assigned by the Human Genome Organisation Committee; this is illustrated in figure 1 which shows a small part of the section covering the class A G-protein-coupled receptors (GPCRs).
The ‘Target information’ field contains sufficient information to attract the interest of the reader while still providing the key data needed for further investigation online. For example, the PubMed references only show the title of the paper to avoid clutter, with the PMID number being sufficient to locate the full citation and all the other information found in the NCBI database. Entries in bold type relate to marketed drugs, or agents in clinical development. A simple online search is generally sufficient to bring up the drug entry and follow its progress (or decline).
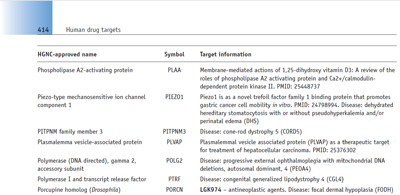
The entries were organised into the different categories that are familiar to drug discovery scientists, namely cell surface receptors, transporters, ion channels and enzymes, the last being presented over two chapters. These target classes are particularly favoured because of their ‘druggability’, but plentiful as they are, they do not make up the majority of the human genome. Some of the myriad proteins involved in cell signalling, membrane turnover and other key functions are also potential drug targets, albeit less straightforward to categorise. For simplicity, entries of this type have been organised into groups based on their subcellular location. Figure 2 shows an example from ‘Internal membranes and organelles’. Of note is that several of the ‘Target information’ entries listed here have disease associations as listed in the online OMIM database (3).
Future-proofing
In a fast moving field like drug target discovery entries will inevitably become outdated, but this is not a problem because the HGNC symbol acts as a stable point of reference, which in most cases remains consistent over time. Although some symbols are discontinued, or open reading frames assigned to a functional gene class as new biological information becomes available, these changes are easily tracked. The current status of a piece of target information is readily checked by searching online. The PubMed search results are particularly useful in that related entries to the initial query often provide more recent information; this may of course support or undermine the evidence supporting the entry as a potential drug target. For example, if the 2012 PMID entry for succinate receptor 1 (last line of figure 1) is entered into the NCBI database, the full citation is displayed along with a more recent 2016 publication supporting the earlier suggestion that SUCRN1 may be novel target for metabolic conditions.
So how many targets are there?
The answer to this question is impossible to predict, not least because some proteins that were once seen as highly unlikely targets for reasons of druggability etc have actually turned out to be unexpectedly tractable. So with potential targets numbering in the thousands, there is no shortage of opportunity and plenty of challenges in turning potential into practical reality.
References
- Human Genome Nomenclature Committee http://www.genenames.org/
- Hjørland B. (2011) Journal of Information Science. 37(5): 546-550
- OMIM database http://www.ncbi.nlm.nih.gov/omim
Where to buy
This book is available to buy here.
|
Biography
|
Related topics
Drug Targets, Molecular Targets, Target Validation