New technique improves the selectivity of kinase inhibitors
Posted: 11 September 2015 | Victoria White
The new technique could cut the potential for side effects and help usher kinase inhibitors past strict regulatory hurdles…

Protein kinase inhibitors have become a powerful weapon in the fight against various life-threatening diseases, including certain types of leukaemia, lung cancer, kidney cancer and squamous cell cancer of the head and neck.
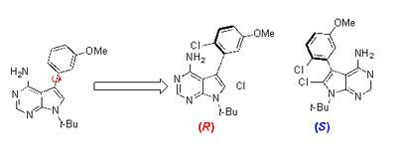

The left-most molecule shows an unstable atropisomer, its axis of chirality shown by the red arrow. The two on the right are stable “arrangements”. CREDIT: Jeffrey Gustafson
However, these drugs often inhibit many different targets, which can lead to side effects and complications in therapeutic use. Now, San Diego State University chemist Jeffrey Gustafson has identified a new technique for improving the selectivity of these drugs and possibly decreasing unwanted side effects in the future.
As the name suggests, kinase inhibitors interrupt the function of kinases and effectively shut down the activity of proteins that contribute to cancer.
“Kinase inhibition has been a watershed for cancer treatment,” said Gustafson. “However, it’s really hard to inhibit a single kinase. The majority of compounds identified inhibit not just one but many kinases, and that can lead to a number of side effects.”
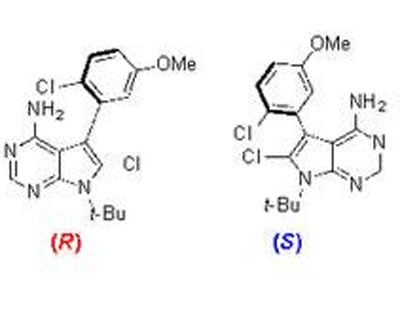
Many of these drug candidates possess examples of a phenomenon known as atropisomerism. Atropisomerism is a form of chirality that arises when the spatial arrangement of molecules in a candidate has a rotatable bond called an axis of chirality. Some axes of chirality are rigid, many kinase inhibitors possess axes of chirality that are freely spinning. The problem is that because you can’t control which “arrangement” of the molecule is present at a given time, the unwanted version could have unintended consequences.
In practice, this means that when medicinal chemists discover a promising kinase inhibitor that exists as two interchanging arrangements, they actually have two different inhibitors. Each one can have quite different biological effects, and it’s difficult to know which version of the molecule actually targets the right protein.
“I think this has really been under-recognised in the field,” Gustafson said. “The field needs strategies to weed out these side effects.”
Discovery could help usher kinase inhibitors past strict regulatory hurdles
Gustafson and his colleagues synthesised atropisomeric compounds known to target a particular family of kinases known as tyrosine kinases. To some of these compounds, the researchers added a single chlorine atom which effectively served as a brake to keep the atropisomer from spinning around, locking the molecule into either a right-handed or a left-handed version.
When the researchers screened both the modified and unmodified versions against their target kinases, they found major differences in which kinases the different versions inhibited. The unmodified compound was like a shotgun blast, inhibiting a broad range of kinases. But the locked-in right-handed and left-handed versions were choosier.
“Just by locking them into one or another atropisomeric configuration, not only were they more selective, but they inhibited different kinases,” Gustafson explained.
If drug makers incorporated this technique into their early drug discovery process, he said, it would help identify which version of an atropisomeric compound actually targets the kinase they want to target, cutting the potential for side effects and helping to usher drugs past strict regulatory hurdles.
Related topics
Drug Discovery, Kinases
Related organisations
San Diego State University


