Improving the translation of disease biology though phenomics
Posted: 20 July 2018 | Paul Andrews (National Screening Centre - Scientific Lead - Dundee Laboratory) | No comments yet
How times change. Up to 15 years ago, you would be hard-pressed to find a drug discovery conference with a track dedicated to phenotypic approaches. The underpinning science was called high-content imaging or analysis, being mainly confined to academic drug discovery labs and a handful of pioneers in industry. but overall this was given little attention and often dismissed by the chemistry-dominated leadership in the industry…


Today any self-respecting pharma-focused conference agenda isn’t complete without at least one session on the use of more physiological phenotypic assays for drug discovery. Now it’s all about 3D models and multi-parametric data, iPSC models and patient-centric approaches.
The driver for this shift was a dawning of realisation, triggered by the much-cited 2011 review by David Swinney and John Anthony entitled “How were new medicines made”.1 The title was subtly innocent but the conclusions were full of impact. Retrospective analysis of FDA-approvals showed that more empirical, phenotypic, approaches where very good at finding first-in-class drugs, especially given the level of investment in this area. However, this new era of enlightenment we are now experiencing was most-likely precipitated by the stark impact of economic failure, rather anything else. There was (and probably still is) a profound and persistent inefficiency in the drug discovery and development process – there is a greater than 90% attrition rate, mainly due to lack of efficacy and safety, leading to extraordinarily high costs and an ever-decreasing return on investment, compounded the extended timelines between initial concept and drug approval. A major contributing factor in this failure is that in the rush to drug huge numbers of biochemical targets – borne from the availability of recombinant DNA technology and eventually whole genome sequences – the need for accurate target validation and predictive in vitro cellular models, was overlooked.
The re-emergence of the phenotypic, or phenomics, an approach in drug discovery has thus been driven by economic necessity coupled to a retrospective realisation that phenotypic approaches are more successful and were, in fact, the way almost all drugs were identified. Despite great leaps in our understanding of a raft of fundamental processes in biology, we still struggle when it comes to understanding complexity and variation, in establishing the causative relationships between molecular function and cellular phenotype and in the modelling of dynamic, complex, systems in human disease. The reductionist approach that relies on perturbing single molecular targets in vitro often fails to deliver when the same drugs are tested in the context of tissues, organs and living breathing humans. Renewed interest in phenotypic methods is not just so pharma can re-live the heady days when drug discovery was successful and lower cost, it is because there has been an explosion in the availability of new biological tools, such as better cellular models, models for tissues, including 3D spheroids and more complex organoids, as well as precision genome editing to recapitulate genetic lesions or tag endogenous loci.
Biological advances are also benefitting from new imaging technologies, single-cell transcriptomics and computational methods including machine learning and Artificial Intelligence (AI), thoroughly reviewed elsewhere.2 Moreover, there is an ever-increasing list of powerful approaches to understanding mechanism-of-action and identifying the targets underlying observed phenotypes which allows for ‘rational’ drug design and avoidance of liabilities. The new field of phenomics is still young and there is much to be done. The translational chain still has weak links – not many parts of the pre-clinical discovery phase are humanised, still being largely a patient-material-free zone, relying on animal models of disease and toxicology, which often correlate poorly with human outcomes. It seems like the time has come for high-throughput phenotypic biology to take centre stage in the hunt for novel drugs that suffer from fewer late-stage failures.
So, if the phenotype is the catalyst, then where is the substrate? There is a deep well of academic and clinical biology in the community that remains untapped, for want of translational direction. At the National Phenotypic Screening Centre (NPSC) we identified this as an opportunity and in 2016 launched an industrial consortium framework called the Phenomics Discovery Initiative (PDi) that has been successfully sourcing the best biology and developing and screening phenotypic assays that tend toward the more complex end of the assay spectrum (Figure 1).
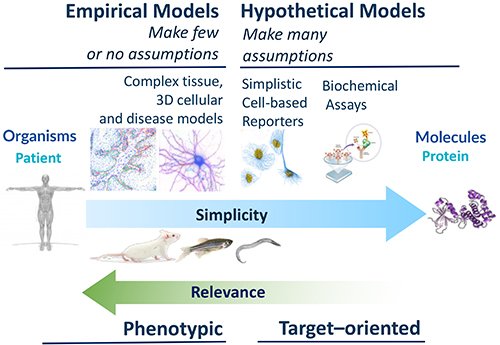

In the infectious diseases and virology space, the lab is developing a lower throughput but highly complex air-liquid interface model of the human bronchus, which has additional applications in other therapeutic areas, a higher throughput, high-content human bronchial epithelial cell assay and a cell-based assay focussing on the replication of enteroviruses linked to mild and severe respiratory diseases. Other ongoing assays focus on the targeting the inflammasome in neurological conditions (bipolar disorder and dementia) and endoplasmic reticulum (ER) stress in an iPSC-derived hepatocyte disease model. We find there is no shortage of proposals put forward in our crowd-sourcing calls.
Phenotypic assay development and screening requires a special blend of skills, in addition to core competency in automation and liquid handling. Ideally, cell biologists take a central role whilst working closely with screening scientists, image and data analysts, engineers and programmers, computer scientists and statisticians and not least, chemists. Cell biology knowledge is of paramount importance as developing the most patho / physiologically-relevant assay possible is often non-trivial – sometimes it may be sufficient to have a cell model growing as a homogenous monolayer but in other circumstances, 3D spheroids or multicellular co-culture models, or even organoids are more appropriate. Some assays involve cancer lines that grow readily in a simple medium, other cell types are highly specialised with peculiar media and growth factor requirements.
Some of the biggest challenges are ones that require expertise in maintaining pluripotent ESCs/iPSCs and adult stem cell and then high-level expertise in differentiating them over extended culture times. Complex co-culture assays are another challenge, though these assays promise to be more predictive and hence of high value. Increasingly, cells are being used that have undergone genome engineering using CRISPR/Cas9 – either to introduce a tag into a gene or to reproduce a genetic lesion.
At NPSC, around half of our assays are image-based and ‘high content’ whilst the other half uses flow cytometry. Microscopes, when set up correctly, are quantitative measuring devices that can acquire images that reveal multiscale spatio-temporal information. High content analysis (HCA) is especially powerful because it unlocks an extensive and rich source of information about subcellular behaviour, single cells, groups of cells and tissues that describes their properties and responses to intrinsic cues and extrinsic perturbations and can give in-built opportunities for triage – for example by measuring cytotoxicity or undesirable phenotypes. When HCA is applied to good biological assays this phenomics approach represents a powerful and highly-informative means to allow data-driven medicinal chemistry in the early phases drug development process, that can deliver more information, earlier, for medchem and drug metabolism and pharmacokinetics (DMPK) decision-making. Phenomics is also powerful because it can extract multiscale and multiplexed data (eg, probing heterogeneous cell-level behaviour and subcellular processes, or even single molecular targets, simultaneously) and can quantitatively measure endpoints or live events on timescales ranging from milliseconds to several days.
A phenotype can be molecular or pathway-centric – examining the changes in specific proteins – or more holistic, in probing a multitude of readouts of cell physiology. Both approaches have value. Historically, most of the high-content screens were relatively low in information content using one or two parameters, which is not as informative and risks bias towards a narrow hypothesis at the expense of understanding the wider effects that a compound has on cell behaviour. In contrast, a much more powerful approach is to extract hundreds or thousands of parameters from each experimental condition or compound-treated well and build a phenotypic fingerprint or “phenoprint” that describes the complex responses cells have when exposed to different perturbing agents. The significance of many of these parameters may not be comprehensible to the biologist, but nonetheless act as signatures of underlying alterations in biology. By assembling data on the way drugs or chemical tool compounds of known target specificity perturb phenoprints, we can use guilt-by-association using clustering and human visualisation or increasingly by using computational methods such as machine learning and neural networks to computationally infer the mode-of-action of hits from chemical library screens. Indeed, a remarkable recent study3 demonstrated machine learning can predict the activities of compounds used in hundreds of biochemical assays from a single image-based screen. The confluence of biology and technology means the future of phenotypic drug discovery promises to be an exciting one.
Biography


References
- Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011 Jun 24;10(7):507-19.
- Horvath P, Aulner N, Bickle M, Davies AM, Nery ED, Ebner D, Montoya MC, Östling P, Pietiäinen V, Price LS, Shorte SL, Turcatti G, von Schantz C, Carragher NO. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. 2016 Nov;15(11):751-769.
- Simm J, Klambauer G, Arany A, Steijaert M, Wegner JK, Gustin E, Chupakhin V, Chong YT, Vialard J, Buijnsters P, Velter I, Vapirev A, Singh S, Carpenter AE,
- Wuyts R, Hochreiter S, Moreau Y, Ceulemans H. Repurposing High-Throughput Image Assays Enables Biological Activity Prediction for Drug Discovery. Cell Chem Biol. 2018 May 17;25(5):611-618
The rest of this content is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- quarterly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Related topics
Analytical techniques, Drug Discovery, High Throughput Screening (HTS), Imaging



